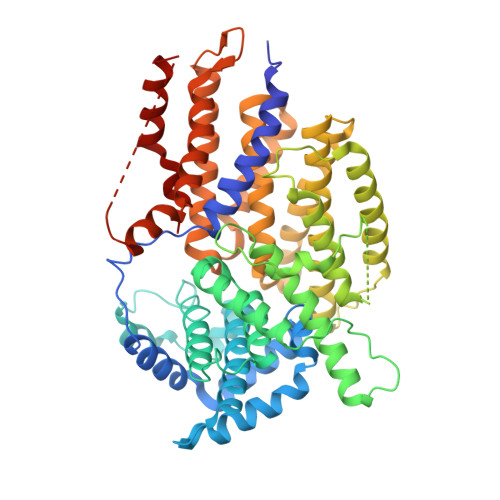
Crystal structure of the yeast inner kinetochore subunit Cep3p.
Bellizzi, J.J., Sorger, P.K., Harrison, S.C.(2007) Structure 15: 1422-1430
- PubMed: 17997968
- DOI: https://doi.org/10.1016/j.str.2007.09.008
- Primary Citation of Related Structures:
2QUQ - PubMed Abstract:
In budding yeast, the four-protein CBF3 complex (Skp1p-Ctf13p-Cep3p-Ndc10p) initiates kinetochore assembly by binding to the CDEIII locus of centromeric DNA. A Cep3p dimer recruits a Skp1p-Ctf13p heterodimer and contacts two sites on CDEIII. We report here the crystal structure, determined at 2.8 A resolution by multiple isomorphous replacement with anomalous scattering, of a truncated Cep3p (Cep3p [47-608]), comprising all but an N-terminal, Zn(2)Cys(6)-cluster, DNA-binding module. Cep3p has a well-ordered structure throughout essentially all of its polypeptide chain, unlike most yeast transcription factors, including those with Zn(2)Cys(6) clusters, such as Gal4p. This difference may reflect an underlying functional distinction: whereas any particular transcription factor must adapt to a variety of upstream activating sites, Cep3p scaffolds kinetochore assembly on centromeres uniformly configured on all 16 yeast chromosomes. We have, using the structure of Cep3p (47-608) and the known structures of Zn(2)Cys(6)-cluster domains, modeled the interaction of Cep3p with CDEIII.
- Jack and Eileen Connors Laboratory of Structural Biology, Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: