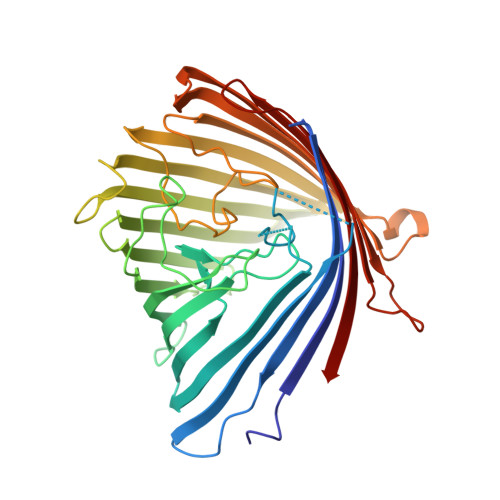
Crystal Structure of the Outer Membrane Protein OpdK from Pseudomonas aeruginosa.
Biswas, S., Mohammad, M.M., Movileanu, L., van den Berg, B.(2008) Structure 16: 1027-1035
- PubMed: 18611376
- DOI: https://doi.org/10.1016/j.str.2008.04.009
- Primary Citation of Related Structures:
2QTK - PubMed Abstract:
In Gram-negative bacteria that do not have porins, most water-soluble and small molecules are taken up by substrate-specific channels belonging to the OprD family. We report here the X-ray crystal structure of OpdK, an OprD family member implicated in the uptake of vanillate and related small aromatic acids. The OpdK structure reveals a monomeric, 18-stranded beta barrel with a kidney-shaped central pore. The OpdK pore constriction is relatively wide for a substrate-specific channel (approximately 8 A diameter), and it is lined by a positively charged patch of arginine residues on one side and an electronegative pocket on the opposite side-features likely to be important for substrate selection. Single-channel electrical recordings of OpdK show binding of vanillate to the channel, and they suggest that OpdK forms labile trimers in the outer membrane. Comparison of the OpdK structure with that of Pseudomonas aeruginosa OprD provides the first qualitative insights into the different substrate specificities of these closely related channels.
- Program in Molecular Medicine, University of Massachusetts Medical School, Worcester, MA 01605, USA.
Organizational Affiliation: