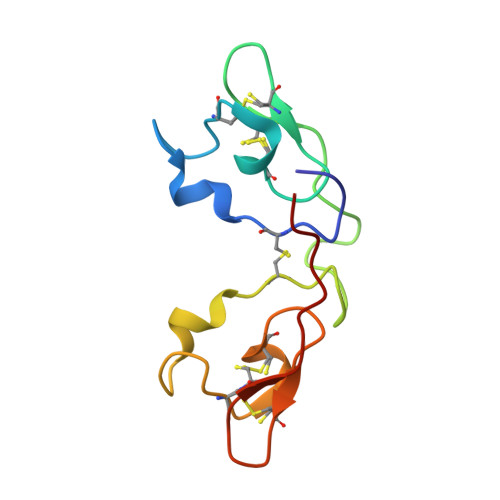
Atomic-resolution crystal structure of the antiviral lectin scytovirin.
Moulaei, T., Botos, I., Ziolkowska, N.E., Bokesch, H.R., Krumpe, L.R., McKee, T.C., O'Keefe, B.R., Dauter, Z., Wlodawer, A.(2007) Protein Sci 16: 2756-2760
- PubMed: 17965185
- DOI: https://doi.org/10.1110/ps.073157507
- Primary Citation of Related Structures:
2QSK, 2QT4 - PubMed Abstract:
The crystal structures of the natural and recombinant antiviral lectin scytovirin (SVN) were solved by single-wavelength anomalous scattering and refined with data extending to 1.3 A and 1.0 A resolution, respectively. A molecule of SVN consists of a single chain 95 amino acids long, with an almost perfect sequence repeat that creates two very similar domains (RMS deviation 0.25 A for 40 pairs of Calpha atoms). The crystal structure differs significantly from a previously published NMR structure of the same protein, with the RMS deviations calculated separately for the N- and C-terminal domains of 5.3 A and 3.7 A, respectively, and a very different relationship between the two domains. In addition, the disulfide bonding pattern of the crystal structures differs from that described in the previously published mass spectrometry and NMR studies.
- Protein Structure Section, Macromolecular Crystallography Laboratory, National Cancer Institute, NCI-Frederick, Frederick, Maryland 21702-1201, USA.
Organizational Affiliation: