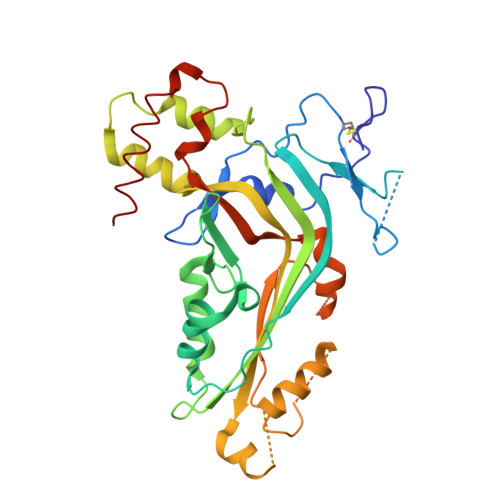
Structure of C8alpha-MACPF reveals mechanism of membrane attack in complement immune defense.
Hadders, M.A., Beringer, D.X., Gros, P.(2007) Science 317: 1552-1554
- PubMed: 17872444
- DOI: https://doi.org/10.1126/science.1147103
- Primary Citation of Related Structures:
2QQH - PubMed Abstract:
Membrane attack is important for mammalian immune defense against invading microorganisms and infected host cells. Proteins of the complement membrane attack complex (MAC) and the protein perforin share a common MACPF domain that is responsible for membrane insertion and pore formation. We determined the crystal structure of the MACPF domain of complement component C8alpha at 2.5 angstrom resolution and show that it is structurally homologous to the bacterial, pore-forming, cholesterol-dependent cytolysins. The structure displays two regions that (in the bacterial cytolysins) refold into transmembrane beta hairpins, forming the lining of a barrel pore. Local hydrophobicity explains why C8alpha is the first complement protein to insert into the membrane. The size of the MACPF domain is consistent with known C9 pore sizes. These data imply that these mammalian and bacterial cytolytic proteins share a common mechanism of membrane insertion.
- Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Department of Chemistry, Faculty of Science, Utrecht University, Padualaan 8, 3584 CH Utrecht, Netherlands.
Organizational Affiliation: