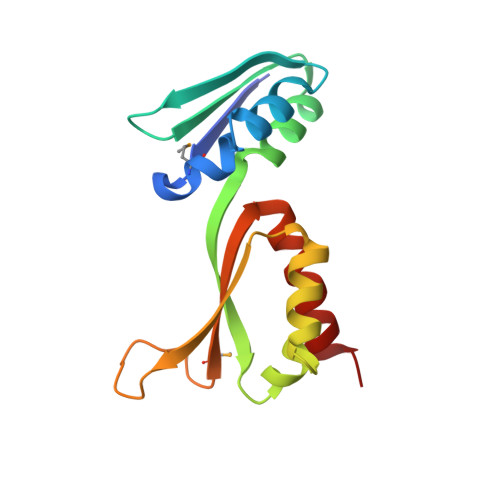
Fragile X mental retardation syndrome: structure of the KH1-KH2 domains of fragile X mental retardation protein.
Valverde, R., Pozdnyakova, I., Kajander, T., Venkatraman, J., Regan, L.(2007) Structure 15: 1090-1098
- PubMed: 17850748
- DOI: https://doi.org/10.1016/j.str.2007.06.022
- Primary Citation of Related Structures:
2QND - PubMed Abstract:
Fragile X syndrome is the most common form of inherited mental retardation in humans, with an estimated prevalence of about 1 in 4000 males. Although several observations indicate that the absence of functional Fragile X Mental Retardation Protein (FMRP) is the underlying basis of Fragile X syndrome, the structure and function of FMRP are currently unknown. Here, we present an X-ray crystal structure of the tandem KH domains of human FMRP, which reveals the relative orientation of the KH1 and KH2 domains and the location of residue Ile304, whose mutation to Asn is associated with a particularly severe incidence of Fragile X syndrome. We show that the Ile304Asn mutation both perturbs the structure and destabilizes the protein.
- Department of Molecular Biophysics and Biochemistry, Yale University, 266 Whitney Avenue, New Haven, CT 06520, USA.
Organizational Affiliation: