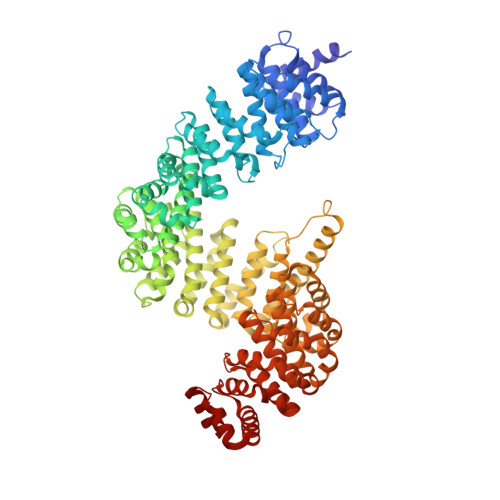
Conformational heterogeneity of karyopherin beta2 is segmental
Cansizoglu, A.E., Chook, Y.M.(2007) Structure 15: 1431-1441
- PubMed: 17997969
- DOI: https://doi.org/10.1016/j.str.2007.09.009
- Primary Citation of Related Structures:
2QMR - PubMed Abstract:
Karyopherinbeta2 (Kap beta2) or transportin imports numerous RNA binding proteins into the nucleus. Kap beta2 binds substrates in the cytoplasm and targets them through the nuclear pore complex, where RanGTP dissociates them in the nucleus. Here we report the 3.0 A crystal structure of unliganded Kap beta2, which consists of a superhelix of 20 HEAT repeats. Together with previously reported structures of NLS and Ran complexes, this structure provides understanding of conformational heterogeneity that accompanies ligand binding. The Kap beta2 superhelix is divided into three major segments. Two of them (HEAT repeats 9-13 and 14-18), which constitute the substrate binding site, are rigid elements that rotate relative to each other about a flexible hinge. The third (HEAT repeats 1-8), which constitutes the Ran binding site, exhibits conformational changes throughout its length. An analogous segmental architecture is also observed in Importin beta, suggesting that it is functionally significant and may be conserved in other import karyopherins.
- Department of Pharmacology, University of Texas Southwestern Medical Center at Dallas, 6001 Forest Park, Dallas, TX 75390-9041, USA.
Organizational Affiliation: