The crystal structure of DNA repair protein RadC from Chlorobium tepidum TLS.
Zhang, R., Duggan, E., Clancy, S., Joachimiak, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA repair protein radC homolog | 126 | Chlorobaculum tepidum TLS | Mutation(s): 0 Gene Names: radC, CT0611 | 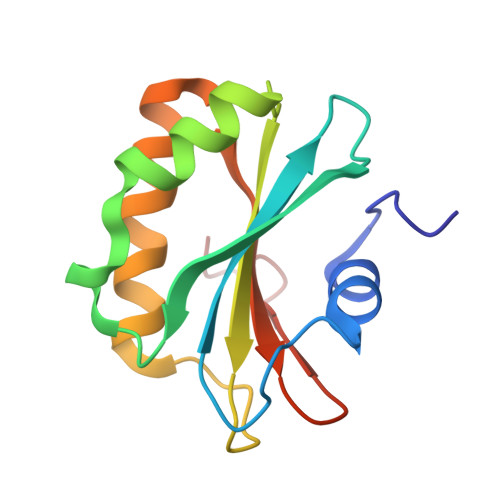 | |
UniProt | |||||
Find proteins for Q8KES1 (Chlorobaculum tepidum (strain ATCC 49652 / DSM 12025 / NBRC 103806 / TLS)) Explore Q8KES1 Go to UniProtKB: Q8KES1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8KES1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 63.434 | α = 90 |
| b = 95.959 | β = 90 |
| c = 192.32 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| SBC-Collect | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| MLPHARE | phasing |