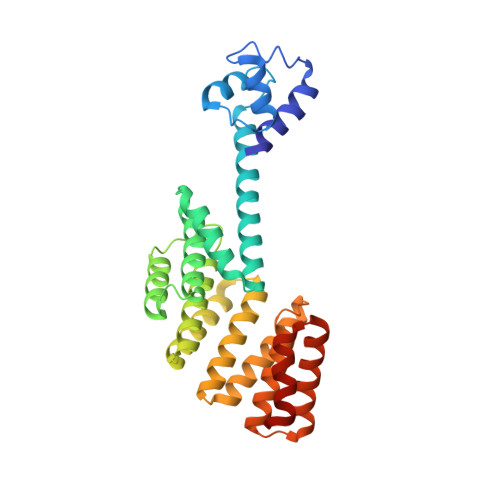
Structure of PlcR: Insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria
Declerck, N., Bouillaut, L., Chaix, D., Rugani, N., Slamti, L., Hoh, F., Lereclus, D., Arold, S.T.(2007) Proc Natl Acad Sci U S A 104: 18490-18495
- PubMed: 17998541
- DOI: https://doi.org/10.1073/pnas.0704501104
- Primary Citation of Related Structures:
2QFC - PubMed Abstract:
Gram-positive bacteria use a wealth of extracellular signaling peptides, so-called autoinducers, to regulate gene expression according to population densities. These "quorum sensing" systems control vital processes such as virulence, sporulation, and gene transfer. Using x-ray analysis, we determined the structure of PlcR, the major virulence regulator of the Bacillus cereus group, and obtained mechanistic insights into the effects of autoinducer binding. Our structural and phylogenetic analysis further suggests that all of those quorum sensors that bind directly to their autoinducer peptide derive from a common ancestor and form a single family (the RNPP family, for Rap/NprR/PlcR/PrgX) with conserved features. As a consequence, fundamentally different processes in different bacterial genera appear regulated by essentially the same autoinducer recognition mechanism. Our results shed light on virulence control by PlcR and elucidate origin and evolution of multicellular behavior in bacteria.
- Institut National de la Santé et de la Recherche Médicale, Unité 554, and Centre National de la Recherche Scientifique, Unité Mixte de Recherche 5048, Centre de Biochimie Structurale, Université de Montpellier, 34090 Montpellier, France.
Organizational Affiliation: