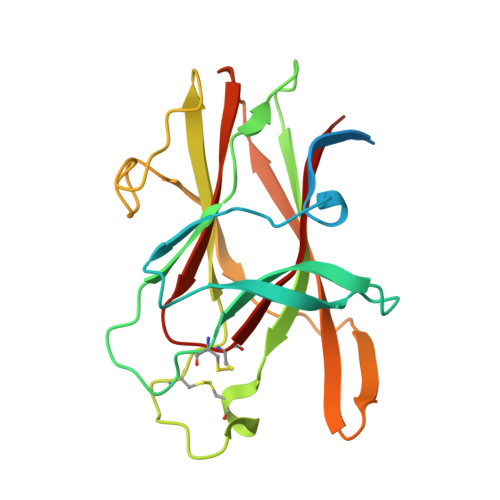
Three-dimensional structure of the EphB2 receptor in complex with an antagonistic peptide reveals a novel mode of inhibition.
Chrencik, J.E., Brooun, A., Recht, M.I., Nicola, G., Davis, L.K., Abagyan, R., Widmer, H., Pasquale, E.B., Kuhn, P.(2007) J Biological Chem 282: 36505-36513
- PubMed: 17897949
- DOI: https://doi.org/10.1074/jbc.M706340200
- Primary Citation of Related Structures:
2QBX - PubMed Abstract:
The Eph family of receptor tyrosine kinases has been implicated in tumorigenesis as well as pathological forms of angiogenesis. Understanding how to modulate the interaction of Eph receptors with their ephrin ligands is therefore of critical interest for the development of therapeutics to treat cancer. Previous work identified a set of 12-mer peptides that displayed moderate binding affinity but high selectivity for the EphB2 receptor. The SNEW antagonistic peptide inhibited the interaction of EphB2 with ephrinB2, with an IC50 of approximately 15 microm. To gain a better molecular understanding of how to inhibit Eph/ephrin binding, we determined the crystal structure of the EphB2 receptor in complex with the SNEW peptide to 2.3-A resolution. The peptide binds in the hydrophobic ligand-binding cleft of the EphB2 receptor, thus competing with the ephrin ligand for receptor binding. However, the binding interactions of the SNEW peptide are markedly different from those described for the TNYL-RAW peptide, which binds to the ligand-binding cleft of EphB4, indicating a novel mode of antagonism. Nevertheless, we identified a conserved structural motif present in all known receptor/ligand interfaces, which may serve as a scaffold for the development of therapeutic leads. The EphB2-SNEW complex crystallized as a homodimer, and the residues involved in the dimerization interface are similar to those implicated in mediating tetramerization of EphB2-ephrinB2 complexes. The structure of EphB2 in complex with the SNEW peptide reveals novel binding determinants that could serve as starting points in the development of compounds that modulate Eph receptor/ephrin interactions and biological activities.
- Department of Cellular Biology and Molecular Biology, The Scripps Research Institute, La Jolla, California 92037, USA.
Organizational Affiliation: