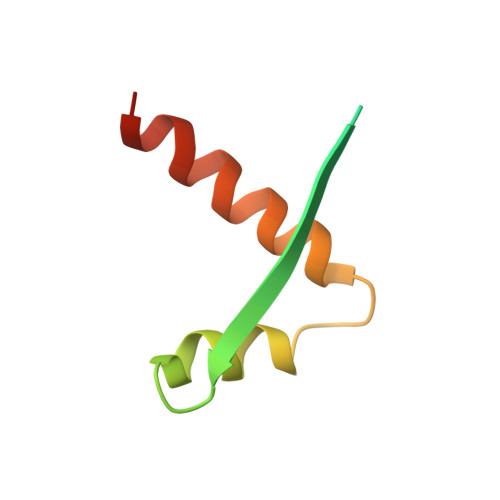
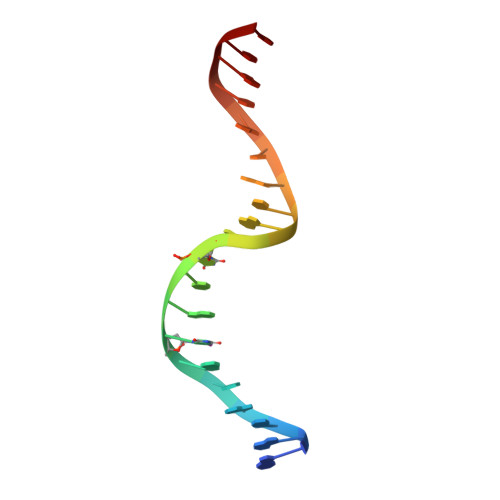
Segrosome structure revealed by a complex of ParR with centromere DNA.
Schumacher, M.A., Glover, T.C., Brzoska, A.J., Jensen, S.O., Dunham, T.D., Skurray, R.A., Firth, N.(2007) Nature 450: 1268-1271
- PubMed: 18097417
- DOI: https://doi.org/10.1038/nature06392
- Primary Citation of Related Structures:
2Q2K - PubMed Abstract:
The stable inheritance of genetic material depends on accurate DNA partition. Plasmids serve as tractable model systems to study DNA segregation because they require only a DNA centromere, a centromere-binding protein and a force-generating ATPase. The centromeres of partition (par) systems typically consist of a tandem arrangement of direct repeats. The best-characterized par system contains a centromere-binding protein called ParR and an ATPase called ParM. In the first step of segregation, multiple ParR proteins interact with the centromere repeats to form a large nucleoprotein complex of unknown structure called the segrosome, which binds ParM filaments. pSK41 ParR binds a centromere consisting of multiple 20-base-pair (bp) tandem repeats to mediate both transcription autoregulation and segregation. Here we report the structure of the pSK41 segrosome revealed in the crystal structure of a ParR-DNA complex. In the crystals, the 20-mer tandem repeats stack pseudo-continuously to generate the full-length centromere with the ribbon-helix-helix (RHH) fold of ParR binding successive DNA repeats as dimer-of-dimers. Remarkably, the dimer-of-dimers assemble in a continuous protein super-helical array, wrapping the DNA about its positive convex surface to form a large segrosome with an open, solenoid-shaped structure, suggesting a mechanism for ParM capture and subsequent plasmid segregation.
- Department of Biochemistry and Molecular Biology, University of Texas, M.D. Anderson Cancer Center, Unit 1000, Houston, TX 77030, USA. maschuma@mdanderson.org
Organizational Affiliation: