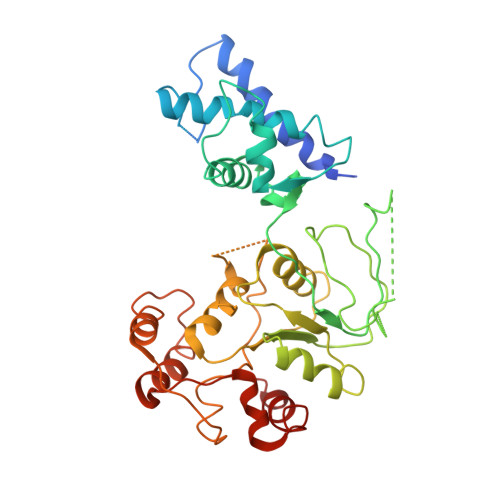
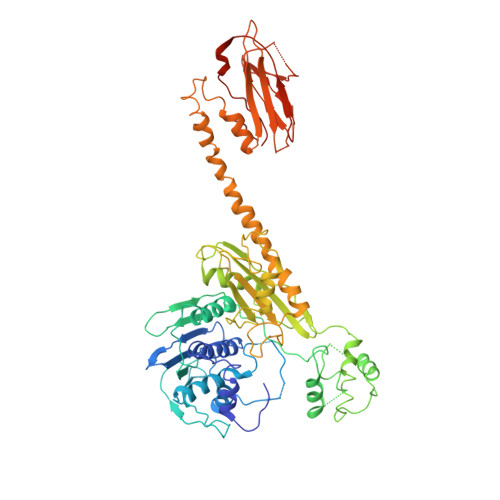
Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI
Corbett, K.D., Benedetti, P., Berger, J.M.(2007) Nat Struct Mol Biol 14: 611-619
- PubMed: 17603498
- DOI: https://doi.org/10.1038/nsmb1264
- Primary Citation of Related Structures:
2Q2E - PubMed Abstract:
Type II topoisomerases help disentangle chromosomes to facilitate cell division. To advance understanding of the structure and dynamics of these essential enzymes, we have determined the crystal structure of an archaeal type IIB topoisomerase, topo VI, at 4.0-A resolution. The 220-kDa heterotetramer adopts a 'twin-gate' architecture, in which a pair of ATPase domains at one end of the enzyme is poised to coordinate DNA movements into the enzyme and through a set of DNA-cleaving domains at the other end. Small-angle X-ray scattering studies show that nucleotide binding elicits a major structural reorganization that is propagated to the enzyme's DNA-cleavage center, explaining how ATP is coupled to DNA capture and strand scission. These data afford important insights into the mechanisms of topo VI and related proteins, including type IIA topoisomerases and the Spo11 meiotic recombination endonuclease.
- Department of Molecular and Cellular Biology, QB3 Institute, Stanley Hall #3220, University of California, Berkeley, California 94720-3220, USA.
Organizational Affiliation: