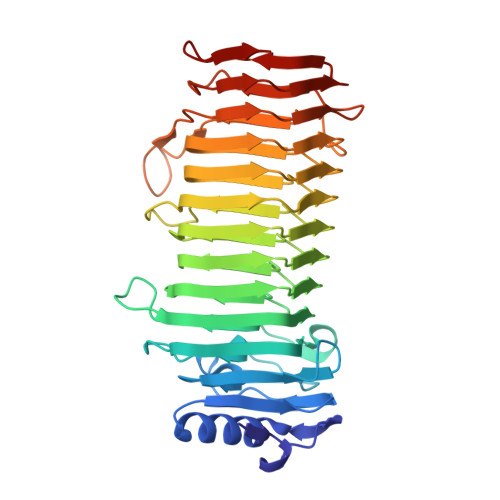
Structural and Mutational Characterization of the Catalytic A-module of the Mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii.
Rozeboom, H.J., Bjerkan, T.M., Kalk, K.H., Ertesvag, H., Holtan, S., Aachmann, F.L., Valla, S., Dijkstra, B.W.(2008) J Biological Chem 283: 23819-23828
- PubMed: 18574239
- DOI: https://doi.org/10.1074/jbc.M804119200
- Primary Citation of Related Structures:
2PYG, 2PYH - PubMed Abstract:
Alginate is a family of linear copolymers of (1-->4)-linked beta-d-mannuronic acid and its C-5 epimer alpha-l-guluronic acid. The polymer is first produced as polymannuronic acid and the guluronic acid residues are then introduced at the polymer level by mannuronan C-5-epimerases. The structure of the catalytic A-module of the Azotobacter vinelandii mannuronan C-5-epimerase AlgE4 has been determined by x-ray crystallography at 2.1-A resolution. AlgE4A folds into a right-handed parallel beta-helix structure originally found in pectate lyase C and subsequently in several polysaccharide lyases and hydrolases. The beta-helix is composed of four parallel beta-sheets, comprising 12 complete turns, and has an amphipathic alpha-helix near the N terminus. The catalytic site is positioned in a positively charged cleft formed by loops extending from the surface encompassing Asp(152), an amino acid previously shown to be important for the reaction. Site-directed mutagenesis further implicates Tyr(149), His(154), and Asp(178) as being essential for activity. Tyr(149) probably acts as the proton acceptor, whereas His(154) is the proton donor in the epimerization reaction.
- Laboratory of Biophysical Chemistry, GBB, University of Groningen, Nijenborgh 4, Groningen, The Netherlands.
Organizational Affiliation: