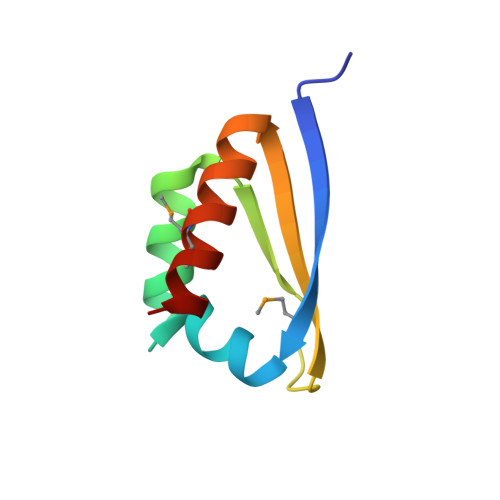
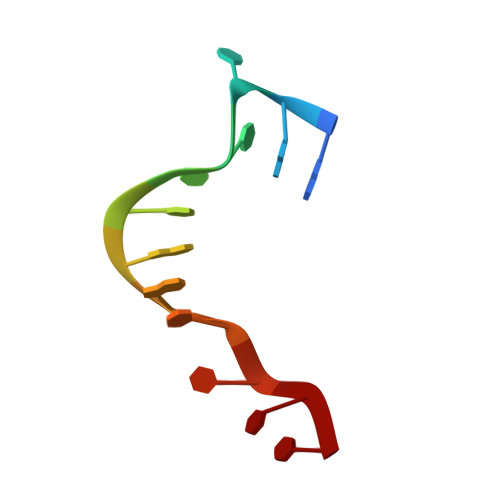
X-ray crystallographic and NMR studies of protein-protein and protein-nucleic acid interactions involving the KH domains from human poly(C)-binding protein-2.
Du, Z., Lee, J.K., Fenn, S., Tjhen, R., Stroud, R.M., James, T.L.(2007) RNA 13: 1043-1051
- PubMed: 17526645
- DOI: https://doi.org/10.1261/rna.410107
- Primary Citation of Related Structures:
2PQU, 2PY9 - PubMed Abstract:
Poly(C)-binding proteins (PCBPs) are KH (hnRNP K homology) domain-containing proteins that recognize poly(C) DNA and RNA sequences in mammalian cells. Binding poly(C) sequences via the KH domains is critical for PCBP functions. To reveal the mechanisms of KH domain-D/RNA recognition and its functional importance, we have determined the crystal structures of PCBP2 KH1 domain in complex with a 12-nucleotide DNA corresponding to two repeats of the human C-rich strand telomeric DNA and its RNA equivalent. The crystal structures reveal molecular details for not only KH1-DNA/RNA interaction but also protein-protein interaction between two KH1 domains. NMR studies on a protein construct containing two KH domains (KH1 + KH2) of PCBP2 indicate that KH1 interacts with KH2 in a way similar to the KH1-KH1 interaction. The crystal structures and NMR data suggest possible ways by which binding certain nucleic acid targets containing tandem poly(C) motifs may induce structural rearrangement of the KH domains in PCBPs; such structural rearrangement may be crucial for some PCBP functions.
- Department of Pharmaceutical Chemistry, University of California at San Francisco, San Francisco, CA 94158-2517, USA.
Organizational Affiliation: