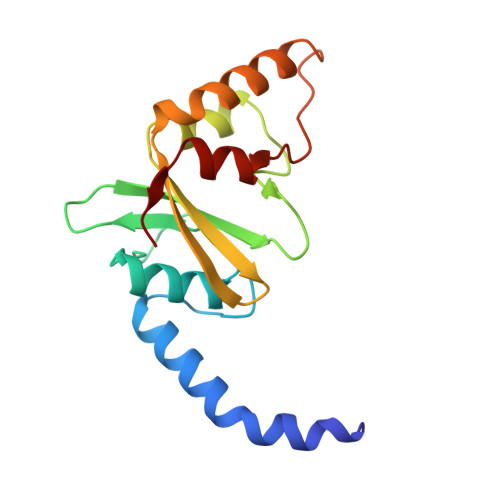
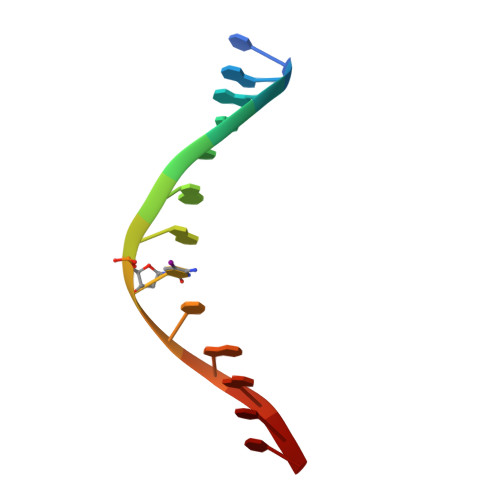
How is modification of the DNA substrate recognized by the PvuII restriction endonuclease?
Horton, J.R., Bonventre, J., Cheng, X.(1998) J Biological Chem 379: 451-458
- PubMed: 9628337
- DOI: https://doi.org/10.1515/bchm.1998.379.4-5.451
- Primary Citation of Related Structures:
2PVI - PubMed Abstract:
In restriction-modification systems, cleavage of substrate sites in cellular DNA by the restriction endonuclease is prevented by the action of a cognate methyltransferase that acts on the same substrate sites. The PvuII restriction endonuclease (R.PvuII) has been structurally characterized in a complex with substrate DNA (Cheng et al., 1994) and as an apoenzyme (Athanasiadis et al., 1994). We report here a structure, determined to 1.9 A resolution by crystallography, of a complex between R.PvuII and iodinated DNA. The presence of an iodine at the 5-carbon of the methylatable cytosine results in the following changes in the protein: His84 moved away from the modified base; this movement was amplified in His85 and disrupts an intersubunit hydrogen bond; and the base modification disturbs the distribution of water molecules that associate with these histidine residues and the area of the scissile bond. Considering these observations, hypotheses are given as to why a similar oligonucleotide, where a methyl group resides on the 5-carbon of the methylatable cytosine, is slowly cleaved by R.PvuII (Rice et al., 1995).
- Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA.
Organizational Affiliation: