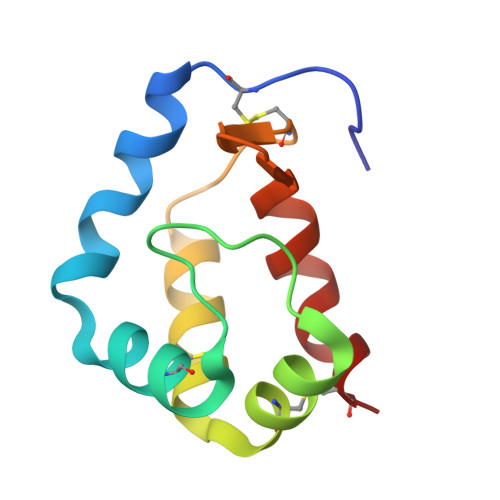
Structure of sylvaticin, a new alpha-elicitin-like protein from Pythium sylvaticum.
Lascombe, M.B., Retailleau, P., Ponchet, M., Industri, B., Blein, J.P., Prange, T.(2007) Acta Crystallogr D Biol Crystallogr 63: 1102-1108
- PubMed: 17881828
- DOI: https://doi.org/10.1107/S0907444907043363
- Primary Citation of Related Structures:
2POS, 2PR0 - PubMed Abstract:
The structure of sylvaticin, a 10 kDa major pythin protein excreted by the parasitic oomycete Pythium sylvaticum, has been determined. Although closely related to alpha-elicitins in its biological response, toxicity and overall structure, sylvaticin presents a number of structural features that make it an unusual member of the elicitin class. Elicitins possess a large hydrophobic cavity and the mechanism of the systemic acquired resistance induced in planta is known to proceed through lipid transport and complexation within this cavity. Unlike other elicitins, sylvaticin contains tryptophan residues, one of which points inwards towards the central cavity, thus limiting access to sterols. In the case of sylvaticin, the sterol-transport mechanism is likely to be of less importance compared with other members of the elicitin family and still remains to be fully characterized.
- Laboratoire de Cristallographie et RMN Biologiques, Université Paris Descartes, UMR 8015 CNRS, Faculté de Pharmacie, 4 Avenue de l'Observatoire, 75006 Paris, France. marie-bernard.lascombe@univ-paris5.fr
Organizational Affiliation: