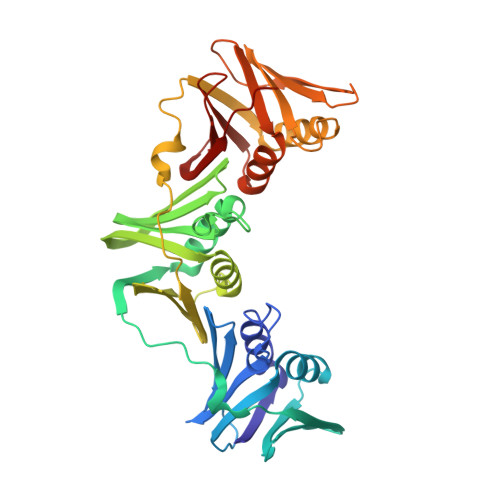
Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp.
Kong, X.P., Onrust, R., O'Donnell, M., Kuriyan, J.(1992) Cell 69: 425-437
- PubMed: 1349852
- DOI: https://doi.org/10.1016/0092-8674(92)90445-i
- Primary Citation of Related Structures:
2POL - PubMed Abstract:
The crystal structure of the beta subunit (processivity factor) of DNA polymerase III holoenzyme has been determined at 2.5 A resolution. A dimer of the beta subunit (M(r) = 2 x 40.6 kd, 2 x 366 amino acid residues) forms a ring-shaped structure lined by 12 alpha helices that can encircle duplex DNA. The structure is highly symmetrical, with each monomer containing three domains of identical topology. The charge distribution and orientation of the helices indicate that the molecule functions by forming a tight clamp that can slide on DNA, as shown biochemically. A potential structural relationship is suggested between the beta subunit and proliferating cell nuclear antigen (PCNA, the eukaryotic polymerase delta [and epsilon] processivity factor), and the gene 45 protein of the bacteriophage T4 DNA polymerase.
- Laboratory of Molecular Biophysics, Rockefeller University, New York, New York 10021.
Organizational Affiliation: