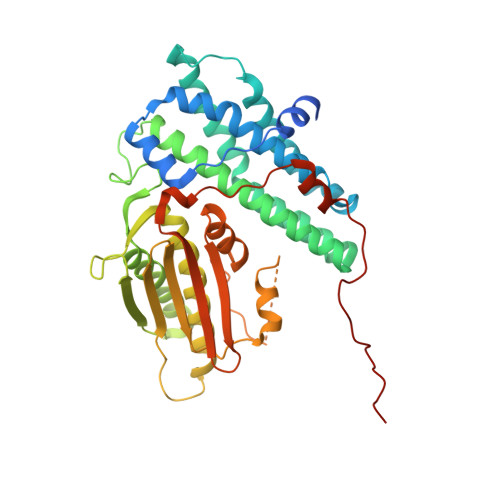
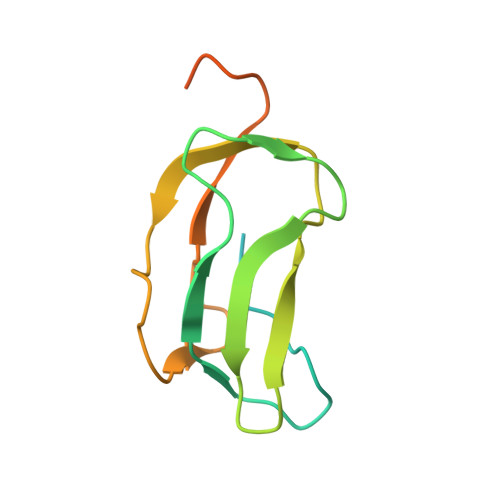
Crystal Structure of an Asymmetric complex of Pyruvate Dehydrogenase Kinase 3 with Lipoyl domain 2 and its Biological Implications
Devedjiev, Y., Steussy, C.N., Vassylyev, D.G.(2007) J Mol Biology 370: 407-416
- PubMed: 17532006
- DOI: https://doi.org/10.1016/j.jmb.2007.04.083
- Primary Citation of Related Structures:
2PNR - PubMed Abstract:
A homodimer of pyruvate dehydrogenase kinase (PDHK) is an integral part of pyruvate dehydrogenase complex (PDC) to which it is anchored primarily through the inner lipoyl-bearing domains (L2) of transacetylase component. The catalytic cycle of PDHK and its translocation over the PDC surface is thought to be mediated by the "symmetric" and "asymmetric" modes, in which the PDHK dimer binds to two and one L2-domain(s), respectively. Whereas the structure of the symmetric PDHK/L2 complex was reported, the structural organization and functional role of the asymmetric complex remain obscure. Here, we report the crystal structure of the asymmetric PDHK3/L2 complex that reveals several functionally important features absent from the previous structures. First, the PDHK3 subunits have distinct conformations: one subunit exhibits "open" and the other "closed" configuration of the putative substrate-binding cleft. Second, access to the closed cleft is additionally restricted by local unwinding of the adjacent alpha-helix. Modeling indicates that the target peptide might gain access to the PDHK active center through the open but not through the closed cleft. Third, the ATP-binding loop in one PDHK3 subunit adopts an open conformation, implying that the nucleotide loading into the active site is mediated by the inactive "pre-insertion" binding mode. Altogether our data suggest that the asymmetric complex represents a physiological state in which binding of a single L2-domain activates one of the PDHK protomers while inactivating another. Thus, the L2-domains likely act not only as the structural anchors but also modulate the catalytic cycle of PDHK.
- Department of Biochemistry and Molecular Genetics, University of Alabama at Birmingham, School of Medicine and Dentistry, Kaul Genetics Building, Birmingham, Al 35294, USA.
Organizational Affiliation: