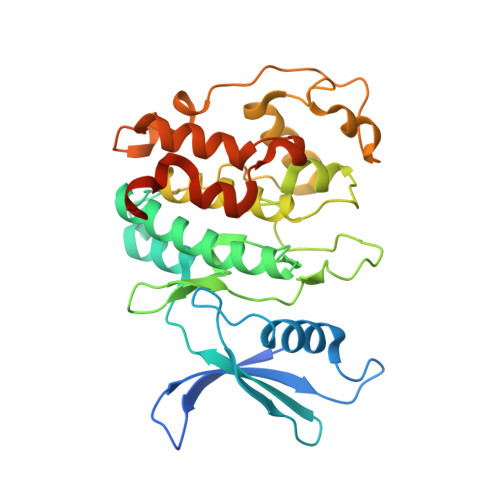
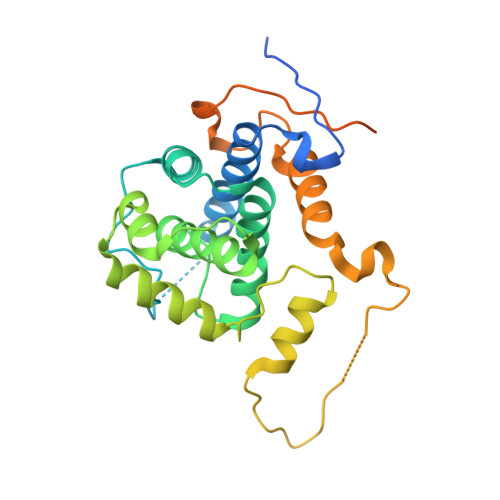
Structure of the Pho85-Pho80 CDK-Cyclin Complex of the Phosphate-Responsive Signal Transduction Pathway
Huang, K., Ferrin-O'Connell, I., Zhang, W., Leonard, G.A., O'Shea, E.K., Quiocho, F.A.(2007) Mol Cell 28: 614-623
- PubMed: 18042456
- DOI: https://doi.org/10.1016/j.molcel.2007.09.013
- Primary Citation of Related Structures:
2PK9, 2PMI - PubMed Abstract:
The ability to sense and respond appropriately to environmental changes is a primary requirement of all living organisms. In response to phosphate limitation, Saccharomyces cerevisiae induces transcription of a set of genes involved in the regulation of phosphate acquisition from the ambient environment. A signal transduction pathway (the PHO pathway) mediates this response, with Pho85-Pho80 playing a vital role. Here we report the X-ray structure of Pho85-Pho80, a prototypic structure of a CDK-cyclin complex functioning in transcriptional regulation in response to environmental changes. The structure revealed a specific salt link between a Pho85 arginine and a Pho80 aspartate that makes phosphorylation of the Pho85 activation loop dispensable and that maintains a Pho80 loop conformation for possible substrate recognition. It further showed two sites on the Pho80 cyclin for high-affinity binding of the transcription factor substrate (Pho4) and the CDK inhibitor (Pho81) that are markedly distant to each other and the active site.
- Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, TX 77030, USA.
Organizational Affiliation: