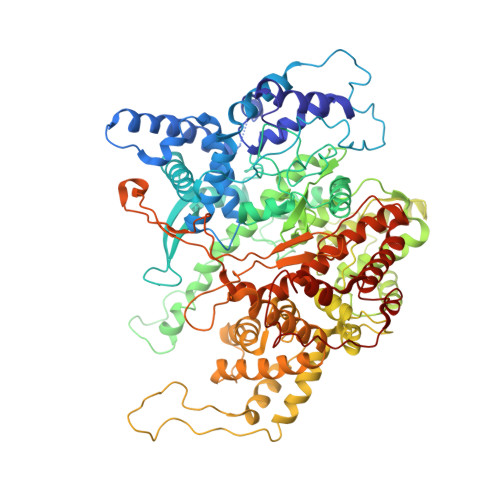
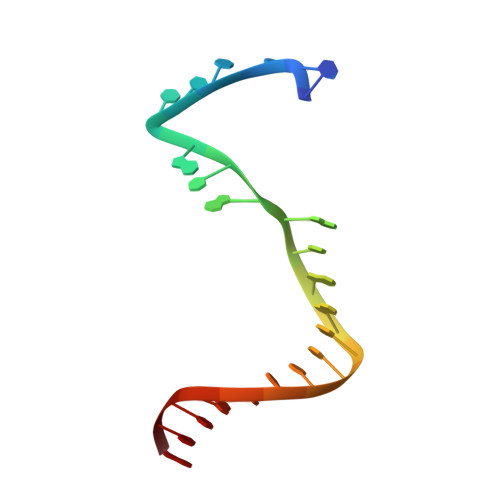
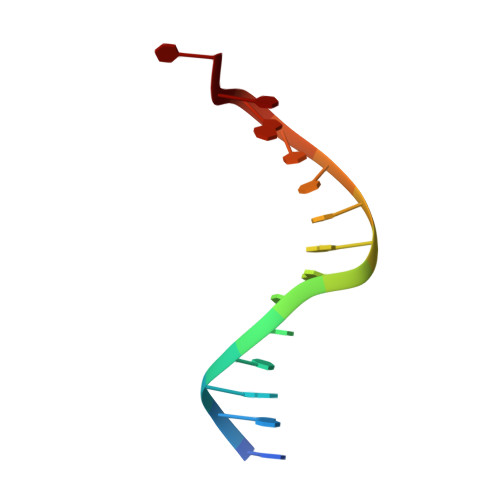
Mechanism for de novo RNA synthesis and initiating nucleotide specificity by t7 RNA polymerase.
Kennedy, W.P., Momand, J.R., Yin, Y.W.(2007) J Mol Biology 370: 256-268
- PubMed: 17512007
- DOI: https://doi.org/10.1016/j.jmb.2007.03.041
- Primary Citation of Related Structures:
2PI4, 2PI5 - PubMed Abstract:
DNA-directed RNA polymerases are capable of initiating synthesis of RNA without primers, the first catalytic stage of initiation is referred to as de novo RNA synthesis. De novo synthesis is a unique phase in the transcription cycle where the RNA polymerase binds two nucleotides rather than a nascent RNA polymer and a single nucleotide. For bacteriophage T7 RNA polymerase, transcription begins with a marked preference for GTP at the +1 and +2 positions. We determined the crystal structures of T7 RNA polymerase complexes captured during the de novo RNA synthesis. The DNA substrates in the structures in the complexes contain a common Phi 10 duplex promoter followed by a unique five base single-stranded extension of template DNA whose sequences varied at positions +1 and +2, thereby allowing for different pairs of initiating nucleotides GTP, ATP, CTP or UTP to bind. The structures show that the initiating nucleotides bind RNA polymerase in locations distinct from those described previously for elongation complexes. Selection bias in favor of GTP as an initiating nucleotide is accomplished by shape complementarity, extensive protein side-chain and strong base-stacking interactions for the guanine moiety in the enzyme active site. Consequently, an initiating GTP provides the largest stabilization force for the open promoter conformation.
- Department of Chemistry and Biochemistry, Institute of Cellular and Molecular Biology, University of Texas at Austin, Austin, TX 78712, USA.
Organizational Affiliation: