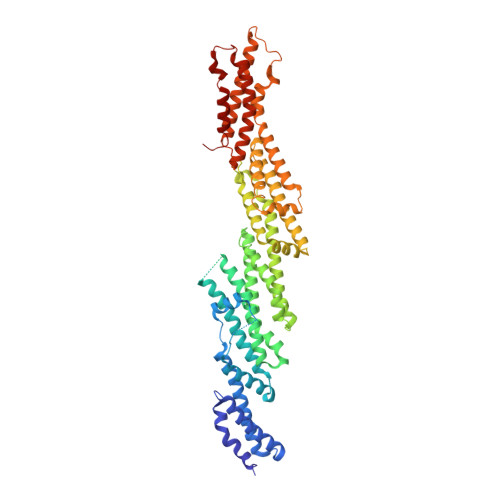The crystal structure of mouse Exo70 reveals unique features of the mammalian exocyst.
Moore, B.A., Robinson, H.H., Xu, Z.(2007) J Mol Biology 371: 410-421
- PubMed: 17583731
- DOI: https://doi.org/10.1016/j.jmb.2007.05.018
- Primary Citation of Related Structures:
2PFT, 2PFV - PubMed Abstract:
The exocyst is a eukaryotic tethering complex necessary for the fusion of exocytic vesicles with the plasma membrane. Its function in vivo is tightly regulated by interactions with multiple small GTPases. Exo70, one of the eight subunits of the exocyst, is important for the localization of the exocyst to the plasma membrane. It interacts with TC10 and Rho3 GTPases in mammals and yeast, respectively, and has been shown recently to bind to the actin-polymerization complex Arp2/3. Here, we present the crystal structure of Mus musculus Exo70 at 2.25 A resolution. Exo70 is composed of alpha-helices in a series of right-handed helix-turn-helix motifs organized into a long rod of length 170 A and width 35 A. Although the alpha-helical organization of this molecule is similar to that in Saccharomyces cerevisiae Exo70, major structural differences are observed on the surface of the molecule, at the domain boundaries, and in various loop structures. In particular, the C-terminal domain of M. musculus Exo70 adopts a new orientation relative to the N-terminal half not seen in S. cerevisiae Exo70 structures. Given the low level of sequence conservation within Exo70, this structure provides new insights into our understanding of many species-specific functions of the exocyst.
- Life Sciences Institute and Department of Biological Chemistry, Medical School, University of Michigan, Ann Arbor, MI 48109, USA.
Organizational Affiliation: