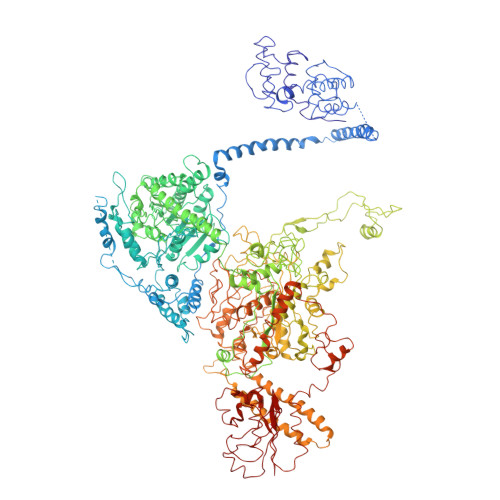
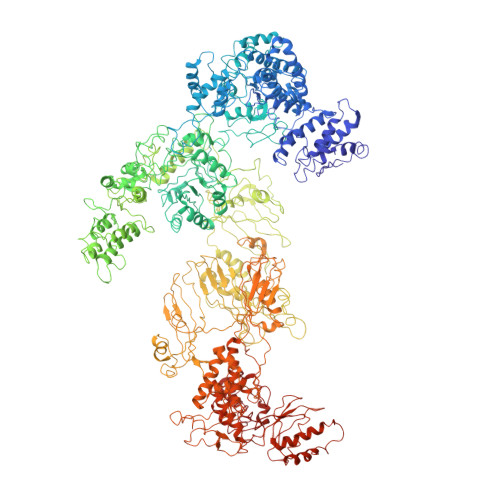
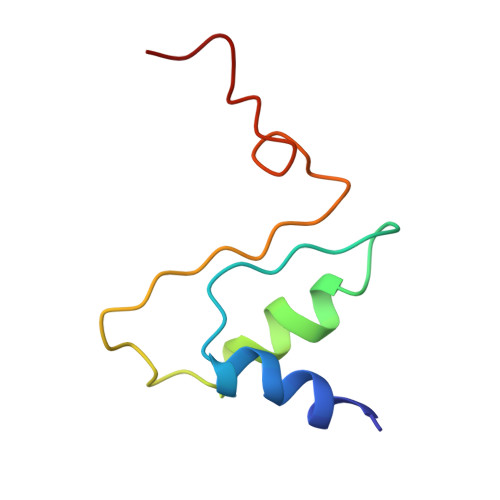
Structural Insights of Yeast Fatty Acid Synthase
Xiong, Y., Lomakin, I.B., Steitz, T.A.(2007) Cell 129: 319-332
- PubMed: 17448991
- DOI: https://doi.org/10.1016/j.cell.2007.03.013
- Primary Citation of Related Structures:
2PFF - PubMed Abstract:
In yeast, the whole metabolic pathway for making 16- and 18-carbon fatty acids is carried out by fatty acid synthase, a 2.6 megadalton molecular-weight macromolecular assembly containing six copies of all eight catalytic centers. We have determined its crystal structure, which illuminates how this enzyme is initially activated and then carries out multiple steps of synthesis in each of six sterically isolated reaction chambers. Six of the catalytic sites are in the wall of the assembly facing an acyl carrier protein (ACP) bound to the ketoacyl synthase domain. Two-dimensional diffusion of substrates to the catalytic sites may be achieved by the electrostatically negative ACP swinging to each of the six electrostatically positive catalytic sites. The phosphopantetheinyl transferase domain lies outside the shell of the assembly, inaccessible to ACP that lies inside, suggesting that the attachment of the pantetheine arm to ACP must occur before complete assembly of the complex.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520-8114, USA.
Organizational Affiliation: