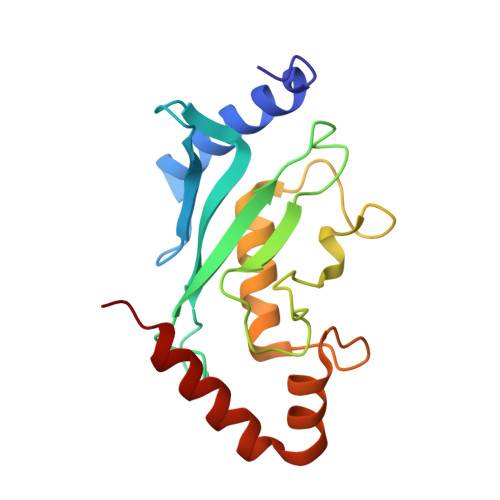
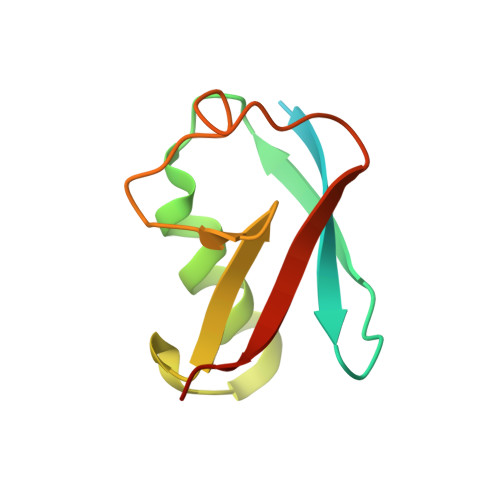
Structure and Analysis of a Complex between SUMO and Ubc9 Illustrates Features of a Conserved E2-Ubl Interaction.
Capili, A.D., Lima, C.D.(2007) J Mol Biology 369: 608-618
- PubMed: 17466333
- DOI: https://doi.org/10.1016/j.jmb.2007.04.006
- Primary Citation of Related Structures:
2PE6 - PubMed Abstract:
The SUMO E2 Ubc9 serves as a lynchpin in the SUMO conjugation pathway, interacting with the SUMO E1 during activation, with thioester linked SUMO after E1 transfer and with the substrate and SUMO E3 ligases during conjugation. Here, we describe the structure determination of a non-covalent complex between human Ubc9 and SUMO-1 at 2.4 A resolution. Non-covalent interactions between Ubc9 and SUMO are conserved in human and yeast insomuch as human Ubc9 interacts with each of the human SUMO isoforms, and yeast Ubc9 interacts with Smt3, the yeast SUMO ortholog. Structural comparisons reveal similarities to several other non-covalent complexes in the ubiquitin pathway, suggesting that the non-covalent Ubc9-SUMO interface may be important for poly-SUMO chain formation, for E2 recruitment to SUMO conjugated substrates, or for mediating E2 interactions with either E1 or E3 ligases. Biochemical analysis suggests that this surface is less important for E1 activation or di-SUMO-2 formation, but more important for E3 interactions and for poly-SUMO chain formation when the chain exceeds more than two SUMO proteins.
- Structural Biology Program, Sloan-Kettering Institute, New York, NY 10021, USA.
Organizational Affiliation: