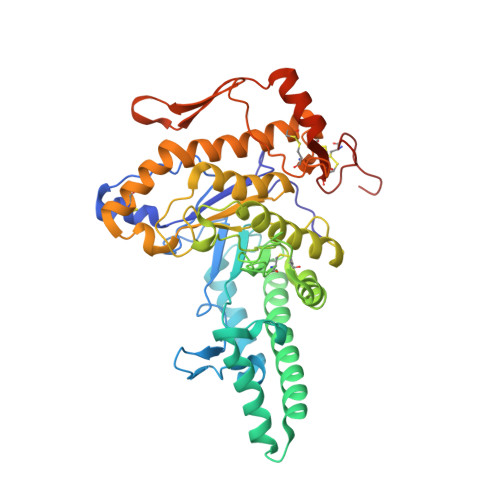
Structure of Human Hyaluronidase-1, a Hyaluronan Hydrolyzing Enzyme Involved in Tumor Growth and Angiogenesis
Chao, K.L., Muthukumar, L., Herzberg, O.(2007) Biochemistry 46: 6911-6920
- PubMed: 17503783
- DOI: https://doi.org/10.1021/bi700382g
- Primary Citation of Related Structures:
2PE4 - PubMed Abstract:
Mammalian hyaluronidases hydrolyze hyaluronan, a polysaccharide of diverse physiological roles found in all tissues and body fluids. In addition to its function in normal cellular hyaluronan turnover, human hyaluronidase-1 is implicated in cancer proliferation, angiogenesis, and inflammatory diseases; its expression is up-regulated in advanced stages of bladder cancer, whereas the expression of the alternative splice-variants is down-regulated. The crystal structure reveals a molecule composed of two closely associated domains: a catalytic domain that adopts a distorted (beta/alpha)8 barrel resembling that of bee venom hyaluronidase, and a novel, EGF-like domain, characteristic of involvement in protein-protein interactions and regulatory processes. The structure shows that the fold of this unique EGF-like domain is intact in four alternative splice-variants, whereas the catalytic domain is likely to be unfolded. Thus, these variants may function by competing with the full-length enzyme for the putative protein partner and regulating enzymatic activity in healthy cells.
- Center for Advanced Research in Biotechnology, University of Maryland Biotechnology Institute, Rockville, Maryland 20850, USA.
Organizational Affiliation: