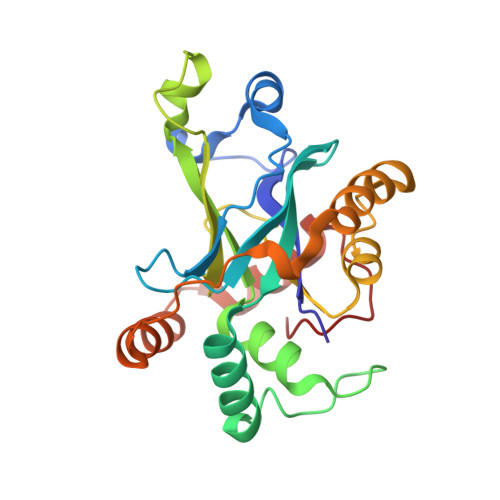
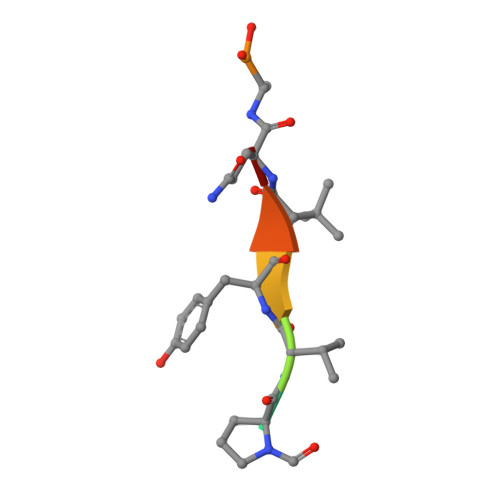
Substrate modulation of enzyme activity in the herpesvirus protease family.
Lazic, A., Goetz, D.H., Nomura, A.M., Marnett, A.B., Craik, C.S.(2007) J Mol Biology 373: 913-923
- PubMed: 17870089
- DOI: https://doi.org/10.1016/j.jmb.2007.07.073
- Primary Citation of Related Structures:
2PBK - PubMed Abstract:
The herpesvirus proteases are an example in which allosteric regulation of an enzyme activity is achieved through the formation of quaternary structure. Here, we report a 1.7 A resolution structure of Kaposi's sarcoma-associated herpesvirus protease in complex with a hexapeptide transition state analogue that stabilizes the dimeric state of the enzyme. Extended substrate binding sites are induced upon peptide binding. In particular, 104 A2 of surface are buried in the newly formed S4 pocket when tyrosine binds at this site. The peptide inhibitor also induces a rearrangement of residues that stabilizes the oxyanion hole and the dimer interface. Concomitant with the structural changes, an increase in catalytic efficiency of the enzyme results upon extended substrate binding. A nearly 20-fold increase in kcat/KM results upon extending the peptide substrate from a tetrapeptide to a hexapeptide exclusively due to a KM effect. This suggests that the mechanism by which herpesvirus proteases achieve their high specificity is by using extended substrates to modulate both the structure and activity of the enzyme.
- Department of Pharmaceutical Chemistry, University of California, San Francisco, CA 94158-2517, USA.
Organizational Affiliation: