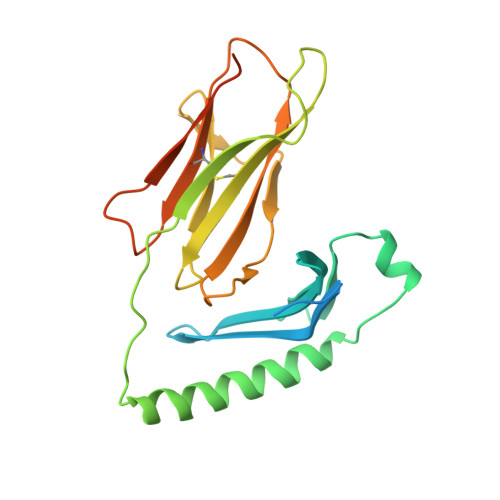
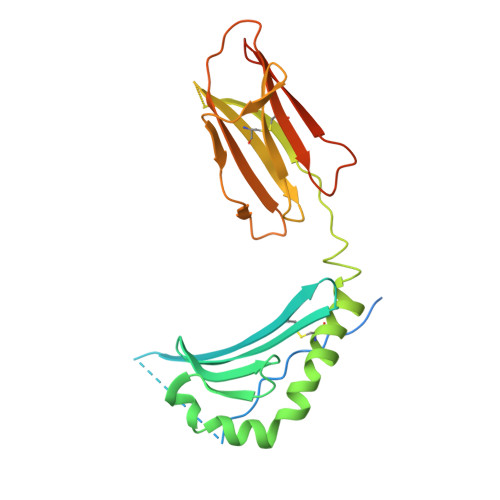
A new twist in TCR diversity revealed by a forbidden alphabeta TCR.
McBeth, C., Seamons, A., Pizarro, J.C., Fleishman, S.J., Baker, D., Kortemme, T., Goverman, J.M., Strong, R.K.(2008) J Mol Biology 375: 1306-1319
- PubMed: 18155234
- DOI: https://doi.org/10.1016/j.jmb.2007.11.020
- Primary Citation of Related Structures:
2P1Y, 2P24 - PubMed Abstract:
We report crystal structures of a negatively selected T cell receptor (TCR) that recognizes two I-A(u)-restricted myelin basic protein peptides and one of its peptide/major histocompatibility complex (pMHC) ligands. Unusual complementarity-determining region (CDR) structural features revealed by our analyses identify a previously unrecognized mechanism by which the highly variable CDR3 regions define ligand specificity. In addition to the pMHC contact residues contributed by CDR3, the CDR3 residues buried deep within the V alpha/V beta interface exert indirect effects on recognition by influencing the V alpha/V beta interdomain angle. This phenomenon represents an additional mechanism for increasing the potential diversity of the TCR repertoire. Both the direct and indirect effects exerted by CDR residues can impact global TCR/MHC docking. Analysis of the available TCR structures in light of these results highlights the significance of the V alpha/V beta interdomain angle in determining specificity and indicates that TCR/pMHC interface features do not distinguish autoimmune from non-autoimmune class II-restricted TCRs.
- Division of Basic Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA.
Organizational Affiliation: