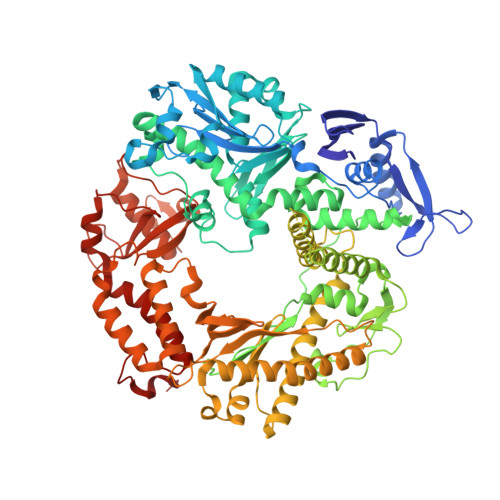
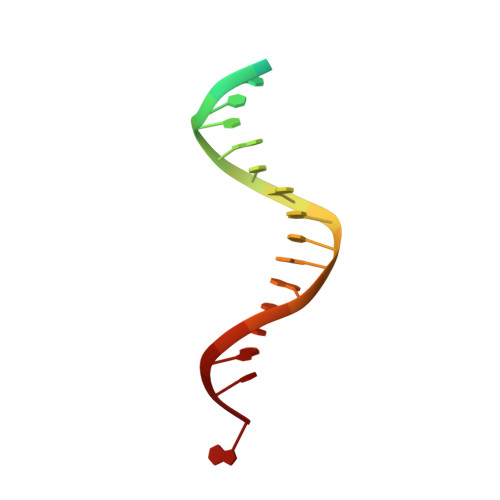
Caught bending the a-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide.
Zahn, K.E., Belrhali, H., Wallace, S.S., Doublie, S.(2007) Biochemistry 46: 10551-10561
- PubMed: 17718515
- DOI: https://doi.org/10.1021/bi7008807
- Primary Citation of Related Structures:
2OYQ, 2OZM, 2OZS, 2P5G - PubMed Abstract:
Damage to DNA involving excision of the nucleobase at the N-glycosidic bond forms abasic sites. If a nucleotide becomes incorporated opposite an unrepaired abasic site during DNA synthesis, most B family polymerases obey the A-rule and preferentially incorporate dAMP without instruction from the template. In addition to being potentially mutagenic, abasic sites provide strong blocks to DNA synthesis. A previous crystal structure of an exonuclease deficient variant of the replicative B family DNA polymerase from bacteriophage RB69 (RB69 gp43 exo-) illustrated these properties, showing that the polymerase failed to translocate the DNA following insertion of dAMP opposite an abasic site. We examine four new structures depicting several steps of translesion DNA synthesis by RB69 gp43 exo-, employing a non-natural purine triphosphate analogue, 5-nitro-1-indolyl-2'-deoxyriboside-5'-triphosphate (5-NITP), that is incorporated more efficiently than dAMP opposite abasic sites. Our structures indicate that a dipole-induced dipole stacking interaction between the 5-nitro group and base 3' to the templating lesion explains the enhanced kinetics of 5-NITP. As with dAMP, the DNA fails to translocate following insertion of 5-NIMP, although distortions at the nascent primer terminus contribute less than previously thought in inducing the stall, given that 5-NIMP preserves relatively undistorted geometry at the insertion site following phosphoryl transfer. An open ternary configuration, novel in B family polymerases, reveals an initial template independent binding of 5-NITP adjacent to the active site of the open polymerase, suggesting that closure of the fingers domain shuttles the nucleotide to the active site while testing the substrate against the template.
- Department of Microbiology and Molecular Genetics, University of Vermont, Burlington, Vermont 05405, USA.
Organizational Affiliation: