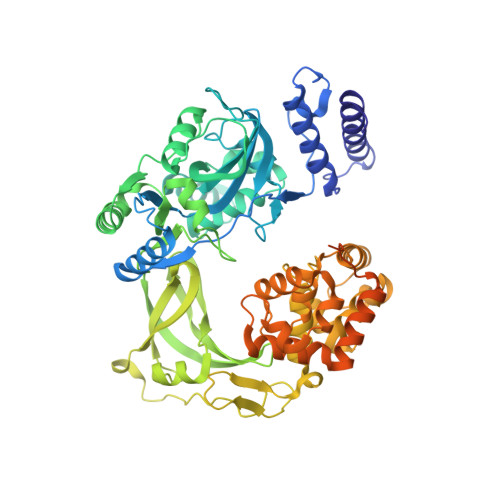
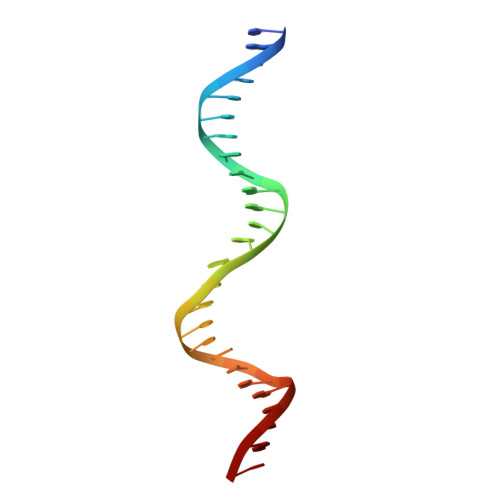
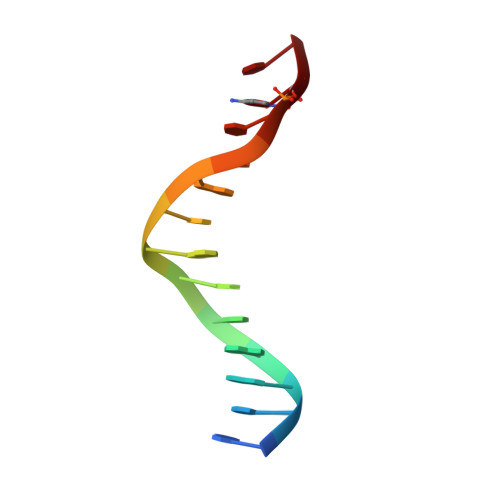
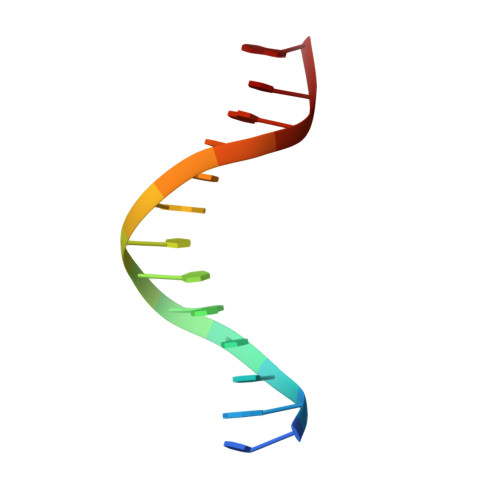
Last Stop on the Road to Repair: Structure of E. coli DNA Ligase Bound to Nicked DNA-Adenylate.
Nandakumar, J., Nair, P.A., Shuman, S.(2007) Mol Cell 26: 257-271
- PubMed: 17466627
- DOI: https://doi.org/10.1016/j.molcel.2007.02.026
- Primary Citation of Related Structures:
2OWO - PubMed Abstract:
NAD(+)-dependent DNA ligases (LigA) are ubiquitous in bacteria and essential for growth. Their distinctive substrate specificity and domain organization vis-a-vis human ATP-dependent ligases make them outstanding targets for anti-infective drug discovery. We report here the 2.3 A crystal structure of Escherichia coli LigA bound to an adenylylated nick, which captures LigA in a state poised for strand closure and reveals the basis for nick recognition. LigA envelopes the DNA within a protein clamp. Large protein domain movements and remodeling of the active site orchestrate progression through the three chemical steps of the ligation reaction. The structure inspires a strategy for inhibitor design.
- Molecular Biology Program, Sloan-Kettering Institute, New York, NY 10021, USA.
Organizational Affiliation: