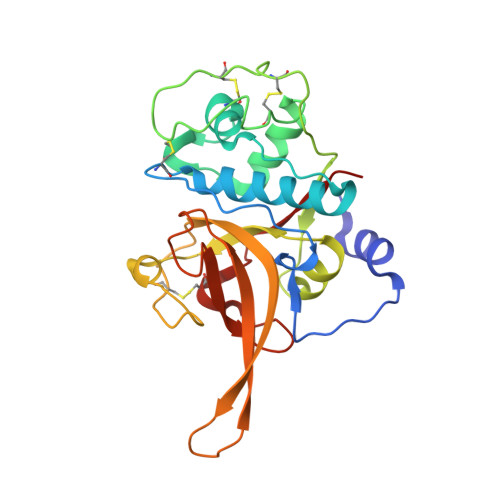
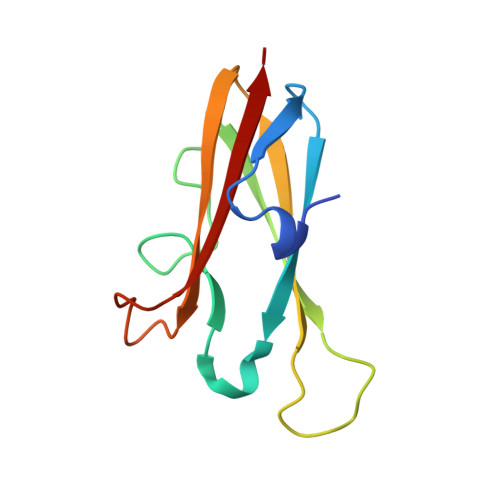
The structure of chagasin in complex with a cysteine protease clarifies the binding mode and evolution of an inhibitor family.
Wang, S.X., Pandey, K.C., Scharfstein, J., Whisstock, J., Huang, R.K., Jacobelli, J., Fletterick, R.J., Rosenthal, P.J., Abrahamson, M., Brinen, L.S., Rossi, A., Sali, A., McKerrow, J.H.(2007) Structure 15: 535-543
- PubMed: 17502099
- DOI: https://doi.org/10.1016/j.str.2007.03.012
- Primary Citation of Related Structures:
2OUL - PubMed Abstract:
Protein inhibitors of proteolytic enzymes regulate proteolysis and prevent the pathological effects of excess endogenous or exogenous proteases. Cysteine proteases are a large family of enzymes found throughout the plant and animal kingdoms. Disturbance of the equilibrium between cysteine proteases and natural inhibitors is a key event in the pathogenesis of cancer, rheumatoid arthritis, osteoporosis, and emphysema. A family (I42) of cysteine protease inhibitors (http://merops.sanger.ac.uk) was discovered in protozoan parasites and recently found widely distributed in prokaryotes and eukaryotes. We report the 2.2 A crystal structure of the signature member of the I42 family, chagasin, in complex with a cysteine protease. Chagasin has a unique variant of the immunoglobulin fold with homology to human CD8alpha. Interactions of chagasin with a target protease are reminiscent of the cystatin family inhibitors. Protein inhibitors of cysteine proteases may have evolved more than once on nonhomologous scaffolds.
- Department of Pathology, University of California, San Francisco, San Francisco, CA 94143, USA.
Organizational Affiliation: