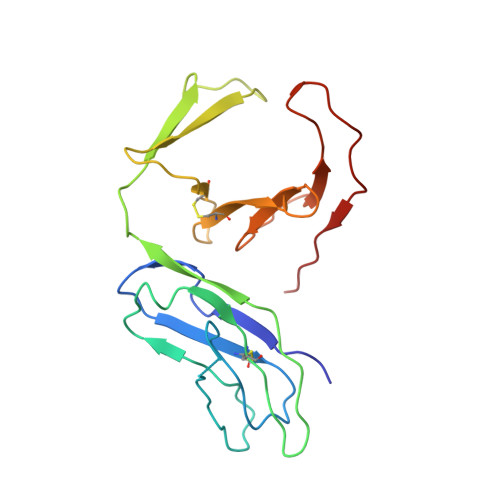
Crystal Structure of 3D Domain Swapped Dimer of Immunoglobulin-Like Transcript 1 (ILT1/LIR7/LILRA2), Molecular Insight into Group 1 Activating Receptor Forming Unique MW Interaction Pattern
Chen, Y., Gao, F., Peng, H., Liu, Y., Gao, G.F.To be published.