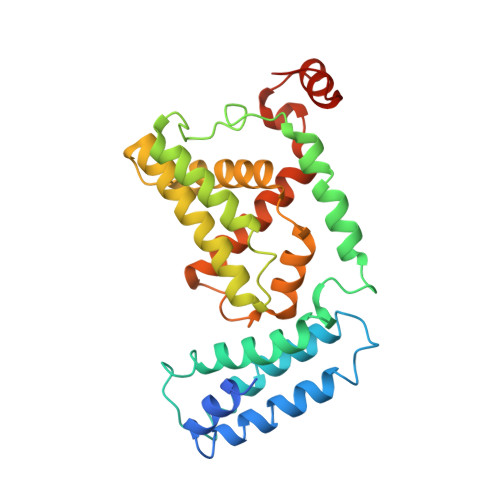
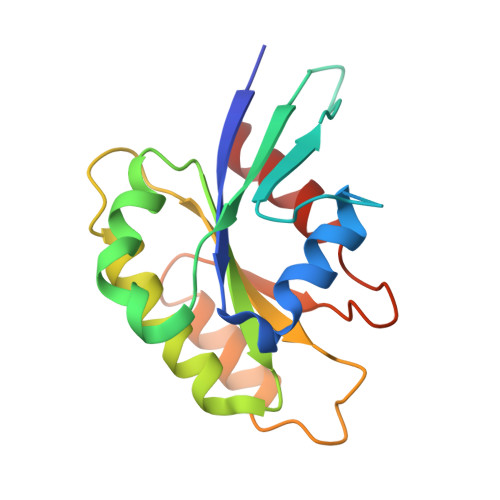
Structural basis for Rab GTPase activation by VPS9 domain exchange factors.
Delprato, A., Lambright, D.G.(2007) Nat Struct Mol Biol 14: 406-412
- PubMed: 17450153
- DOI: https://doi.org/10.1038/nsmb1232
- Primary Citation of Related Structures:
2OT3 - PubMed Abstract:
RABEX-5 and other exchange factors with VPS9 domains regulate endocytic trafficking through activation of the Rab family GTPases RAB5, RAB21 and RAB22. Here we report the crystal structure of the RABEX-5 catalytic core in complex with nucleotide-free RAB21, a key intermediate in the exchange reaction pathway. The structure reveals how VPS9 domain exchange factors recognize Rab GTPase substrates, accelerate GDP release and stabilize the nucleotide-free conformation. We further identify an autoinhibitory element in a predicted amphipathic helix located near the C terminus of the VPS9 domain. The autoinhibitory element overlaps with the binding site for the multivalent effector RABAPTIN-5 and potently suppresses the exchange activity of RABEX-5. Autoinhibition can be partially reversed by mutation of conserved residues on the nonpolar face of the predicted amphipathic helix or by assembly of the complex with RABAPTIN-5.
- Program in Molecular Medicine and Department of Biochemistry & Molecular Pharmacology, University of Massachusetts Medical School, Two Biotech, 373 Plantation Street, Worcester, Massachusetts 01605, USA.
Organizational Affiliation: