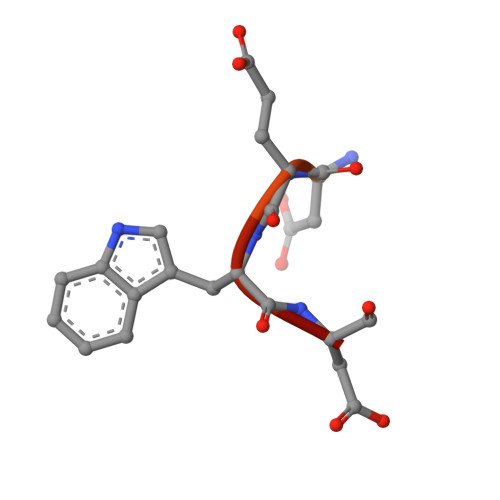
A hydrophobic pocket in the active site of glycolytic aldolase mediates interactions with wiskott-Aldrich syndrome protein.
St-Jean, M., Izard, T., Sygusch, J.(2007) J Biological Chem 282: 14309-14315
- PubMed: 17329259
- DOI: https://doi.org/10.1074/jbc.M611505200
- Primary Citation of Related Structures:
2OT0, 2OT1 - PubMed Abstract:
Aldolase plays essential catalytic roles in glycolysis and gluconeogenesis. However, aldolase is a highly abundant protein that is remarkably promiscuous in its interactions with other cellular proteins. In particular, aldolase binds to highly acidic amino acid sequences, including the C terminus of the Wiskott-Aldrich syndrome protein, an actin nucleation-promoting factor. Here we report the crystal structure of tetrameric rabbit muscle aldolase in complex with a C-terminal peptide of Wiskott-Aldrich syndrome protein. Aldolase recognizes a short, four-residue DEWD motif (residues 498-501), which adopts a loose hairpin turn that folds around the central aromatic residue, enabling its tryptophan side chain to fit into a hydrophobic pocket in the active site of aldolase. The flanking acidic residues in this binding motif provide further interactions with conserved aldolase active site residues Arg-42 and Arg-303, aligning their side chains and forming the sides of the hydrophobic pocket. The binding of Wiskott-Aldrich syndrome protein to aldolase precludes intramolecular interactions of its C terminus with its active site and is competitive with substrate as well as with binding by actin and cortactin. Finally, based on this structure, a novel naphthol phosphate-based inhibitor of aldolase was identified, and its structure in complex with aldolase demonstrated mimicry of the Wiskott-Aldrich syndrome protein-aldolase interaction. The data support a model whereby aldolase exists in distinct forms that regulate glycolysis or actin dynamics.
- Department of Biochemistry, University of Montreal, Montreal, Quebec, Canada.
Organizational Affiliation: