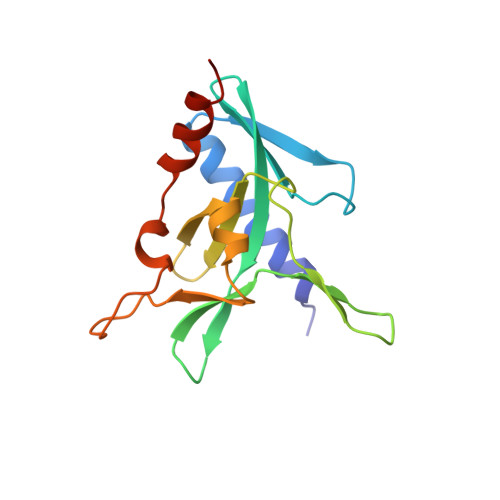
The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif.
Zhao, L., Bonocora, R.P., Shub, D.A., Stoddard, B.L.(2007) EMBO J 26: 2432-2442
- PubMed: 17410205
- DOI: https://doi.org/10.1038/sj.emboj.7601672
- Primary Citation of Related Structures:
2OST - PubMed Abstract:
The homing endonuclease I-Ssp6803I causes the insertion of a group I intron into a bacterial tRNA gene-the only example of an invasive mobile intron within a bacterial genome. Using a computational fold prediction, mutagenic screen and crystal structure determination, we demonstrate that this protein is a tetrameric PD-(D/E)-XK endonuclease - a fold normally used to protect a bacterial genome from invading DNA through the action of restriction endonucleases. I-Ssp6803I uses its tetrameric assembly to promote recognition of a single long target site, whereas restriction endonuclease tetramers facilitate cooperative binding and cleavage of two short sites. The limited use of the PD-(D/E)-XK nucleases by mobile introns stands in contrast to their frequent use of LAGLIDADG and HNH endonucleases - which in turn, are rarely incorporated into restriction/modification systems.
- Graduate Program in Molecular Biophysics, Structure and Design, University of Washington, Seattle, WA, USA.
Organizational Affiliation: