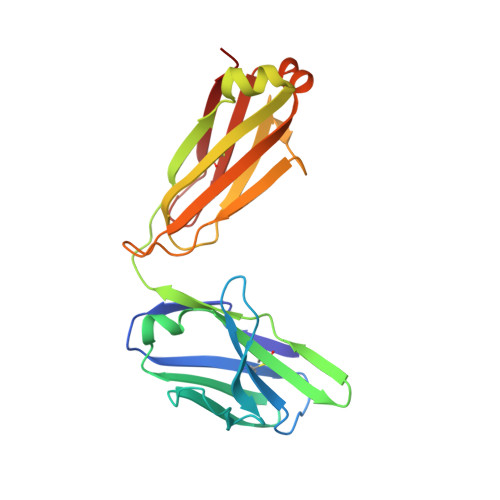
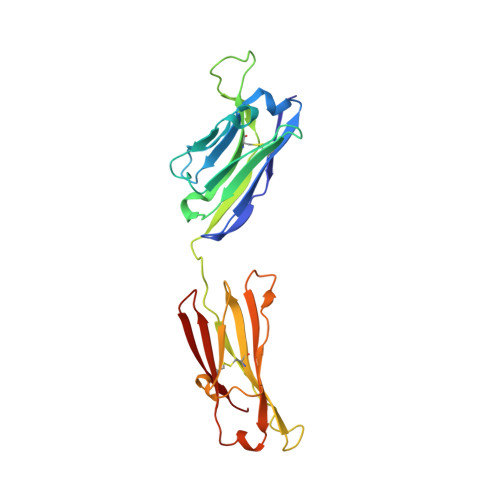
A peptide inhibitor of HIV-1 neutralizing antibody 2G12 is not a structural mimic of the natural carbohydrate epitope on gp120.
Menendez, A., Calarese, D.A., Stanfield, R.L., Chow, K.C., Scanlan, C.N., Kunert, R., Katinger, H., Burton, D.R., Wilson, I.A., Scott, J.K.(2008) FASEB J 22: 1380-1392
- PubMed: 18198210
- DOI: https://doi.org/10.1096/fj.07-8983com
- Primary Citation of Related Structures:
2OQJ - PubMed Abstract:
MAb 2G12 neutralizes HIV-1 by binding with high affinity to a cluster of high-mannose oligosaccharides on the envelope glycoprotein, gp120. Screening of phage-displayed peptide libraries with 2G12 identified peptides that bind specifically, with K(d)s ranging from 0.4 to 200 microM. The crystal structure of a 21-mer peptide ligand in complex with 2G12 Fab was determined at 2.8 A resolution. Comparison of this structure with previous structures of 2G12-carbohydrate complexes revealed striking differences in the mechanism of 2G12 binding to peptide vs. carbohydrate. The peptide occupies a site different from, but adjacent to, the primary carbohydrate-binding site on 2G12, and makes only slightly fewer contacts to the Fab than Man(9)GlcNAc(2) (51 vs. 56, respectively). However, only two antibody contacts with the peptide are hydrogen bonds in contrast to six with Man(9)GlcNAc(2), and only three of the antibody residues that interact with Man(9)GlcNAc(2) also contact the peptide. Thus, this mechanism of peptide binding to 2G12 does not support structural mimicry of the native carbohydrate epitope on gp120, since it neither replicates the oligosaccharide footprint on the antibody nor most of the contact residues. Moreover, 2G12.1 peptide is not an immunogenic mimic of the 2G12 epitope, since antisera produced against it did not bind gp120.
- Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, BC, V5A 1S6, Canada.
Organizational Affiliation: