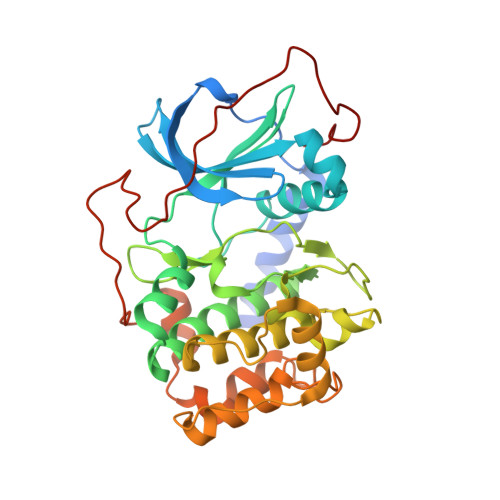
Design and synthesis of pyridine-pyrazolopyridine-based inhibitors of protein kinase B/Akt.
Zhu, G.D., Gong, J., Gandhi, V.B., Woods, K., Luo, Y., Liu, X., Guan, R., Klinghofer, V., Johnson, E.F., Stoll, V.S., Mamo, M., Li, Q., Rosenberg, S.H., Giranda, V.L.(2007) Bioorg Med Chem 15: 2441-2452
- PubMed: 17258463
- DOI: https://doi.org/10.1016/j.bmc.2007.01.010
- Primary Citation of Related Structures:
2OH0, 2OJF - PubMed Abstract:
Thr-211 is one of three different amino acid residues in the kinase domain of protein kinase B/Akt as compared to protein kinase A (PKA), a closely related analog in the same AGC family. In an attempt to improve the potency and selectivity of our indazole-pyridine series of Akt inhibitors over PKA, efforts have focused on the incorporation of a chemical functionality to interact with the hydroxy group of Thr-211. Several substituents including an oxygen anion, amino, and nitro groups have been introduced at the C-6 position of the indazole scaffold, leading to a significant drop in Akt potency. Incorporation of a nitrogen atom into the phenyl ring at the same position (i.e., 9f) maintained the Akt activity and, in some cases, improved the selectivity over PKA. The structure-activity relationships of the new pyridine-pyrazolopyridine series of Akt inhibitors and their structural features when bound to PKA are also discussed.
- Cancer Research, GPRD, Abbott Laboratories, 100 Abbott Park Road, Abbott Park, IL 60064-6101, USA. gui-dong.zhu@abbott.com
Organizational Affiliation: