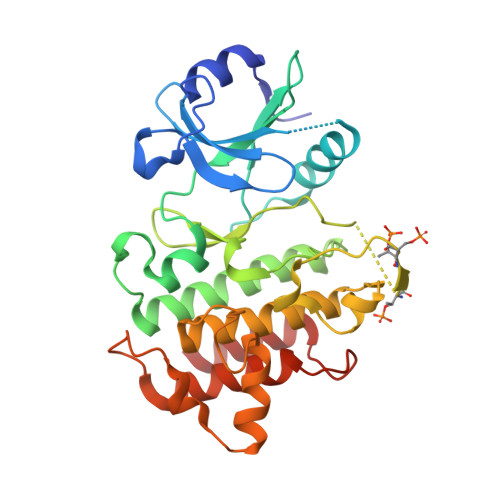
Cutting Edge: IL-1 Receptor-Associated Kinase 4 Structures Reveal Novel Features and Multiple Conformations.
Kuglstatter, A., Villasenor, A.G., Shaw, D., Lee, S.W., Tsing, S., Niu, L., Song, K.W., Barnett, J.W., Browner, M.F.(2007) J Immunol 178: 2641-2645
- PubMed: 17312103
- DOI: https://doi.org/10.4049/jimmunol.178.5.2641
- Primary Citation of Related Structures:
2OIB, 2OIC, 2OID - PubMed Abstract:
IL-1R-associated kinase (IRAK)4 plays a central role in innate and adaptive immunity, and is a crucial component in IL-1/TLR signaling. We have determined the crystal structures of the apo and ligand-bound forms of human IRAK4 kinase domain. These structures reveal several features that provide opportunities for the design of selective IRAK4 inhibitors. The N-terminal lobe of the IRAK4 kinase domain is structurally distinctive due to a loop insertion after an extended N-terminal helix. The gatekeeper residue is a tyrosine, a unique feature of the IRAK family. The IRAK4 structures also provide insights into the regulation of its activity. In the apo structure, two conformations coexist, differing in the relative orientation of the two kinase lobes and the position of helix C. In the presence of an ATP analog only one conformation is observed, indicating that this is the active conformation.
- Roche Palo Alto LLC, 3431 Hillview Avenue, Palo Alto, CA 94304, USA. andreas.kuglstatter@roche.com
Organizational Affiliation: