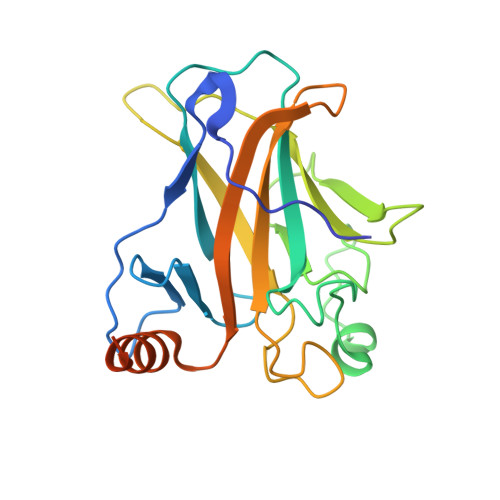
Structure of the human p53 core domain in the absence of DNA.
Wang, Y., Rosengarth, A., Luecke, H.(2007) Acta Crystallogr D Biol Crystallogr 63: 276-281
- PubMed: 17327663
- DOI: https://doi.org/10.1107/S0907444906048499
- Primary Citation of Related Structures:
2OCJ - PubMed Abstract:
The tumor suppressor protein p53 plays a key role in cell-cycle regulation by triggering DNA repair, cell-cycle arrest and apoptosis when the appropriate signal is received. p53 has the classic architecture of a transcription factor, with an amino-terminal transactivation domain, a core DNA-binding domain and carboxy-terminal tetramerization and regulatory domains. The crystal structure of the p53 core domain, which includes the amino acids from residue 96 to residue 289, has been determined in the absence of DNA to a resolution of 2.05 A. Crystals grew in a new monoclinic space group (P2(1)), with unit-cell parameters a = 68.91, b = 69.36, c = 84.18 A, beta = 90.11 degrees . The structure was solved by molecular replacement and has been refined to a final R factor of 20.9% (R(free) = 24.6%). The final model contains four molecules in the asymmetric unit with four zinc ions and 389 water molecules. The non-crystallographic tetramers display different protein contacts from those in other p53 crystals, giving rise to the question of how p53 arranges as a tetramer when it binds its target DNA.
- Department of Molecular Biology and Biochemistry, 3205 McGaugh Hall, University of California, Irvine, CA 92697-3900, USA.
Organizational Affiliation: