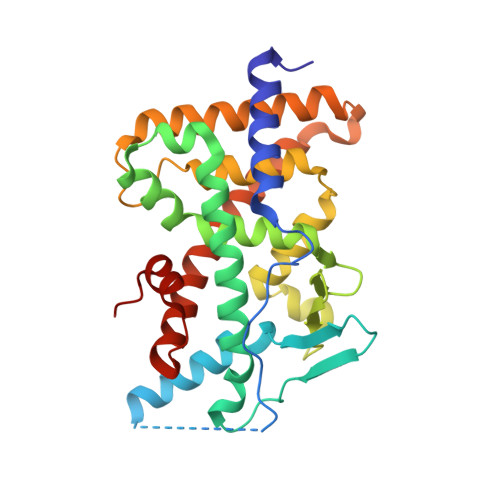
Crystal structure of the PXR-T1317 complex provides a scaffold to examine the potential for receptor antagonism.
Xue, Y., Chao, E., Zuercher, W.J., Willson, T.M., Collins, J.L., Redinbo, M.R.(2007) Bioorg Med Chem 15: 2156-2166
- PubMed: 17215127
- DOI: https://doi.org/10.1016/j.bmc.2006.12.026
- Primary Citation of Related Structures:
2O9I - PubMed Abstract:
The human pregnane X receptor (PXR) recognizes a range of structurally and chemically distinct ligands and plays a key role in regulating the expression of protective gene products involved in the metabolism and excretion of potentially harmful compounds. The identification and development of PXR antagonists is desirable as a potential way to control the up-regulation of drug metabolism pathways during the therapeutic treatment of disease. We present the 2.8A resolution crystal structure of the PXR ligand binding domain (LBD) in complex with T0901317 (T1317), which is also an agonist of another member of the orphan class of the nuclear receptor superfamily, the liver X receptor (LXR). In spite of differences in the size and shape of the receptors' ligand binding pockets, key interactions with this ligand are conserved between human PXR and human LXR. Based on the PXR-T1317 structure, analogues of T1317 were generated with the goal of designing an PXR antagonist effective via the receptor's ligand binding pocket. We find that selectivity in activating PXR versus LXR was achieved; such compounds may be useful in addressing neurodegenerative diseases like Niemann-Pick C. We were not successful, however, in producing a PXR antagonist. Based on these observations, we conclude that the generation of PXR antagonists targeted to the ligand binding pocket may be difficult due to the promiscuity and structural conformability of this xenobiotic sensor.
- Department of Chemistry, CB#3290, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-3290, USA.
Organizational Affiliation: