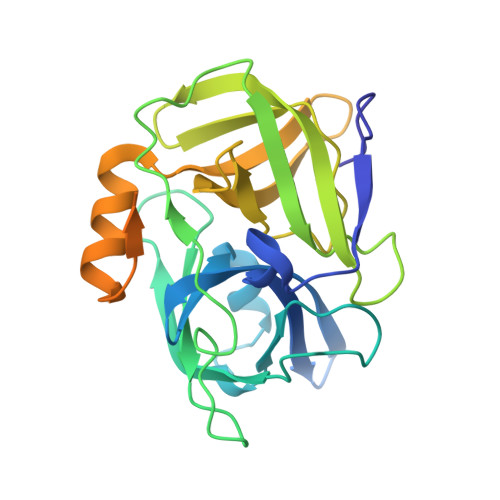
The Structure of a Universally Employed Enazyme: V8 Protease from Staphylococcus Aureus
Prasad, L., Leduc, Y., Hayakawa, K., Delbaere, L.T.J.(2004) Acta Crystallogr D Biol Crystallogr 60: 256-259
- PubMed: 14747701
- DOI: https://doi.org/10.1107/S090744490302599X
- Primary Citation of Related Structures:
1QY6, 2O8L - PubMed Abstract:
V8 protease, an extracellular protease of Staphylococcus aureus, is related to the pancreatic serine proteases. The enzyme cleaves peptide bonds exclusively on the carbonyl side of aspartate and glutamate residues. Unlike the pancreatic serine proteases, V8 protease possesses no disulfide bridges. This is a major evolutionary difference, as all pancreatic proteases have at least two disulfide bridges. The structure of V8 protease shows structural similarity with several other serine proteases, specifically the epidermolytic toxins A and B from S. aureus and trypsin, in which the conformation of the active site is almost identical. V8 protease is also unique in that the positively charged N-terminus is involved in determining the substrate-specificity of the enzyme.
- Department of Biochemistry, University of Saskatchewan, Saskatoon, Saskatchewan S7N 5E5, Canada.
Organizational Affiliation: