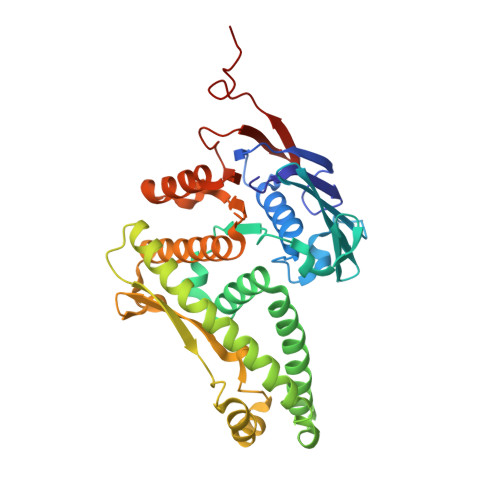
Structural conservation of RecF and Rad50: implications for DNA recognition and RecF function.
Koroleva, O., Makharashvili, N., Courcelle, C.T., Courcelle, J., Korolev, S.(2007) EMBO J 26: 867-877
- PubMed: 17255941
- DOI: https://doi.org/10.1038/sj.emboj.7601537
- Primary Citation of Related Structures:
2O5V - PubMed Abstract:
RecF, together with RecO and RecR, belongs to a ubiquitous group of recombination mediators (RMs) that includes eukaryotic proteins such as Rad52 and BRCA2. RMs help maintain genome stability in the presence of DNA damage by loading RecA-like recombinases and displacing single-stranded DNA-binding proteins. Here, we present the crystal structure of RecF from Deinococcus radiodurans. RecF exhibits a high degree of structural similarity with the head domain of Rad50, but lacks its long coiled-coil region. The structural homology between RecF and Rad50 is extensive, encompassing the ATPase subdomain and the so-called 'Lobe II' subdomain of Rad50. The pronounced structural conservation between bacterial RecF and evolutionarily diverged eukaryotic Rad50 implies a conserved mechanism of DNA binding and recognition of the boundaries of double-stranded DNA regions. The RecF structure, mutagenesis of conserved motifs and ATP-dependent dimerization of RecF are discussed with respect to its role in promoting presynaptic complex formation at DNA damage sites.
- Edward A Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, St Louis, MO 63104, USA.
Organizational Affiliation: