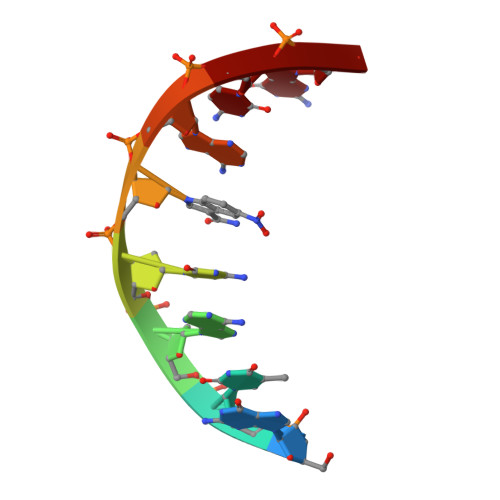
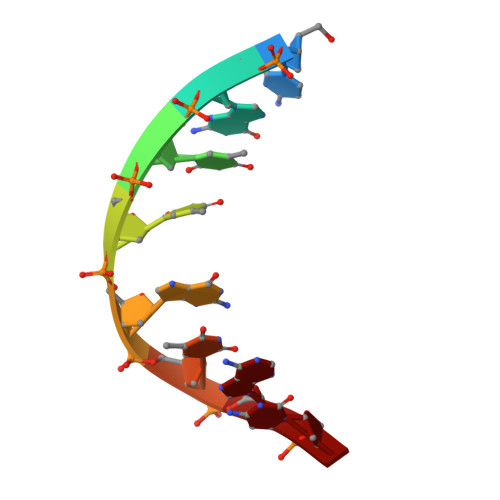
Solution structure and dynamics of DNA duplexes containing the universal base analogues 5-nitroindole and 5-nitroindole 3-carboxamide.
Gallego, J., Loakes, D.(2007) Nucleic Acids Res 35: 2904-2912
- PubMed: 17438041
- DOI: https://doi.org/10.1093/nar/gkm074
- Primary Citation of Related Structures:
2O4Y - PubMed Abstract:
Universal bases hybridize with all other natural DNA or RNA bases, and have applications in PCR and sequencing. We have analysed by nuclear magnetic resonance spectroscopy the structure and dynamics of three DNA oligonucleotides containing the universal base analogues 5-nitroindole and 5-nitroindole-3-carboxamide. In all systems studied, both the 5-nitroindole nucleotide and the opposing nucleotide adopt a standard anti conformation and are fully stacked within the DNA duplex. The 5-nitroindole bases do not base pair with the nucleotide opposite them, but intercalate between this base and an adjacent Watson-Crick pair. In spite of their smooth accommodation within the DNA double-helix, the 5-nitroindole-containing duplexes exist as a dynamic mixture of two different stacking configurations exchanging fast on the chemical shift timescale. These configurations depend on the relative intercalating positions of the universal base and the opposing base, and their exchange implies nucleotide opening motions on the millisecond time range. The structure of these nitroindole-containing duplexes explains the mechanism by which these artificial moieties behave as universal bases.
- Centro de Investigación Príncipe Felipe, Avda. Autopista del Saler 16, 46013 Valencia, Spain. jgallego@cipf.es
Organizational Affiliation: