Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I and Tyr18I
Bateman, K.S., Anderson, S., Lu, W., Qasim, M.A., Huang, K., Laskowski Jr., M., James, M.N.G.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Streptogrisin B | A [auth E] | 185 | Streptomyces griseus | Mutation(s): 0 EC: 3.4.21.81 | 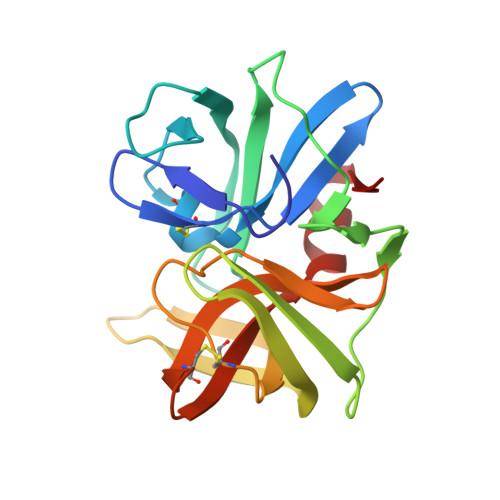 |
UniProt | |||||
Find proteins for P00777 (Streptomyces griseus) Explore P00777 Go to UniProtKB: P00777 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00777 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ovomucoid | B [auth I] | 51 | Meleagris gallopavo | Mutation(s): 1 | 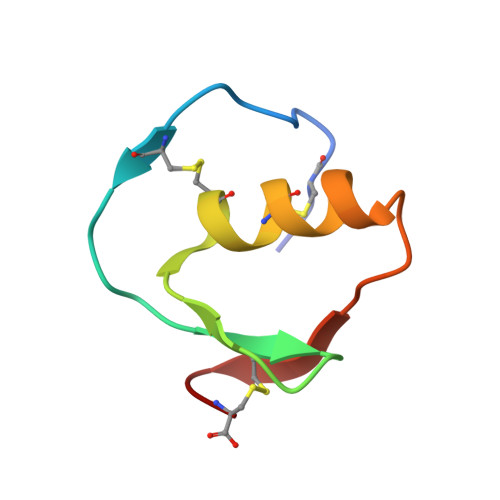 |
UniProt | |||||
Find proteins for P68436 (Meleagris ocellata) Explore P68436 Go to UniProtKB: P68436 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P68436 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 45.44 | α = 90 |
| b = 54.72 | β = 119.01 |
| c = 45.6 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DIP | data collection |
| X-PLOR | model building |
| TNT | refinement |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| X-PLOR | phasing |