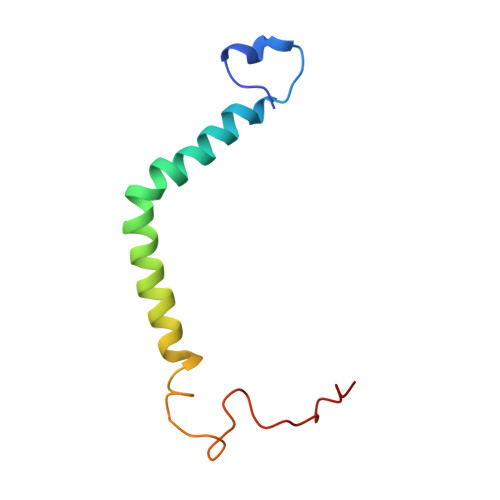
The solution structure of the PufX polypeptide from Rhodobacter sphaeroides.
Tunnicliffe, R.B., Ratcliffe, E.C., Hunter, C.N., Williamson, M.P.(2006) FEBS Lett 580: 6967-6971
- PubMed: 17161397
- DOI: https://doi.org/10.1016/j.febslet.2006.11.065
- Primary Citation of Related Structures:
2ITA, 2NRG - PubMed Abstract:
PufX organizes the photosynthetic reaction centre-light harvesting complex 1 (RC-LH1) core complex of Rhodobacter sphaeroides and facilitates quinol/quinone exchange between the RC and cytochrome bc(1) complexes. The structure of PufX in organic solvent reveals two hydrophobic helices flanked by unstructured termini and connected by a helical bend. The proposed location of basic residues and tryptophans at the membrane interface orients the C-terminal helix along the membrane normal, with the GXXXG motifs in positions unsuitable as direct drivers of dimerisation of the RC-LH1 complex. The N-terminal helix is predicted to extend approximately 40 Anggstrom along the membrane interface.
- Department of Molecular Biology and Biotechnology, University of Sheffield, Firth Court, Western Bank, Sheffield S10 2TN, UK.
Organizational Affiliation: