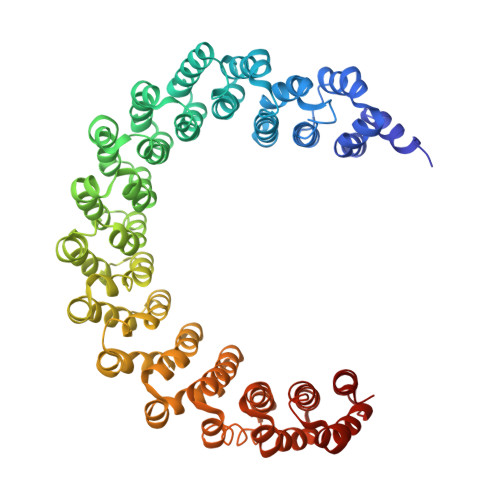Structure of the protein phosphatase 2A holoenzyme
Xu, Y., Xing, Y., Chen, Y., Chao, Y., Lin, Z., Fan, E., Yu, J.W., Strack, S., Jeffrey, P.D., Shi, Y.(2006) Cell 127: 1239-1251
- PubMed: 17174897
- DOI: https://doi.org/10.1016/j.cell.2006.11.033
- Primary Citation of Related Structures:
2NPP, 2NYL, 2NYM - PubMed Abstract:
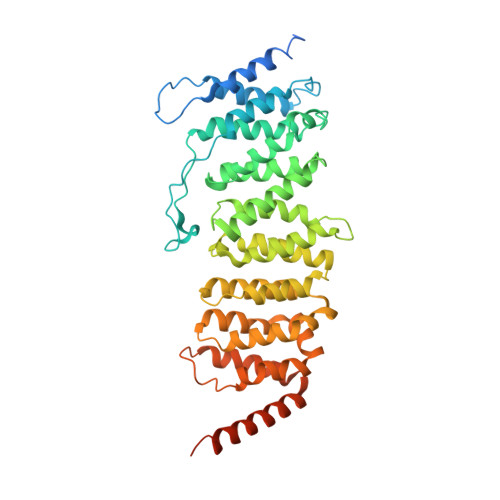
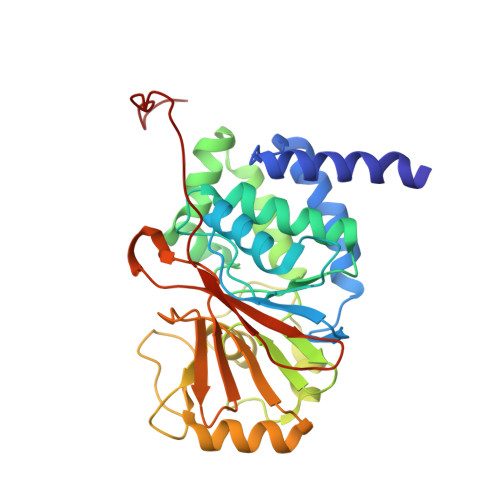
Protein Phosphatase 2A (PP2A) plays an essential role in many aspects of cellular physiology. The PP2A holoenzyme consists of a heterodimeric core enzyme, which comprises a scaffolding subunit and a catalytic subunit, and a variable regulatory subunit. Here we report the crystal structure of the heterotrimeric PP2A holoenzyme involving the regulatory subunit B'/B56/PR61. Surprisingly, the B'/PR61 subunit has a HEAT-like (huntingtin-elongation-A subunit-TOR-like) repeat structure, similar to that of the scaffolding subunit. The regulatory B'/B56/PR61 subunit simultaneously interacts with the catalytic subunit as well as the conserved ridge of the scaffolding subunit. The carboxyterminus of the catalytic subunit recognizes a surface groove at the interface between the B'/B56/PR61 subunit and the scaffolding subunit. Compared to the scaffolding subunit in the PP2A core enzyme, formation of the holoenzyme forces the scaffolding subunit to undergo pronounced conformational rearrangements. This structure reveals significant ramifications for understanding the function and regulation of PP2A.
- Department of Molecular Biology, Lewis Thomas Laboratory, Princeton University, Princeton, NJ 08544, USA.
Organizational Affiliation: