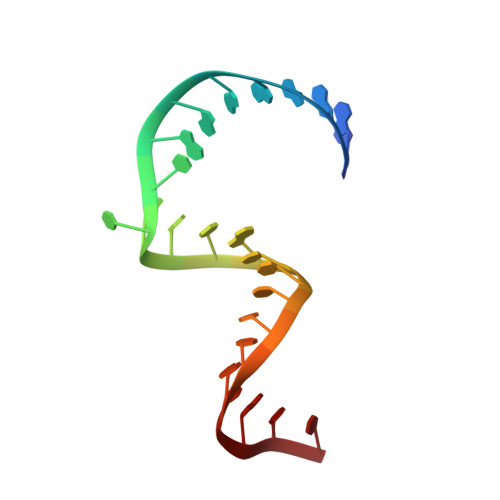
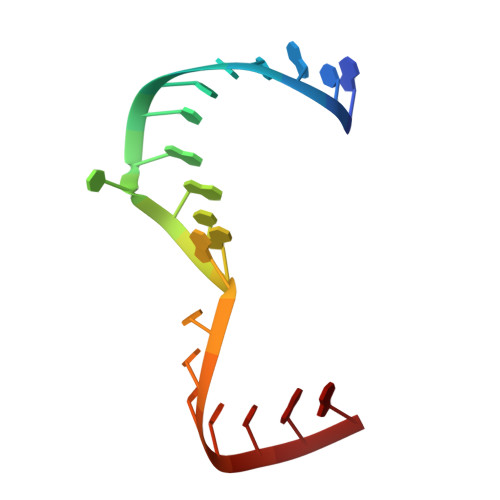Functional architecture of HCV IRES domain II stabilized by divalent metal ions in the crystal and in solution.
Dibrov, S.M., Johnston-Cox, H., Weng, Y.H., Hermann, T.(2007) Angew Chem Int Ed Engl 46: 226-229
- PubMed: 17131443
- DOI: https://doi.org/10.1002/anie.200603807
- Primary Citation of Related Structures:
2NOK - Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093, USA.
Organizational Affiliation: