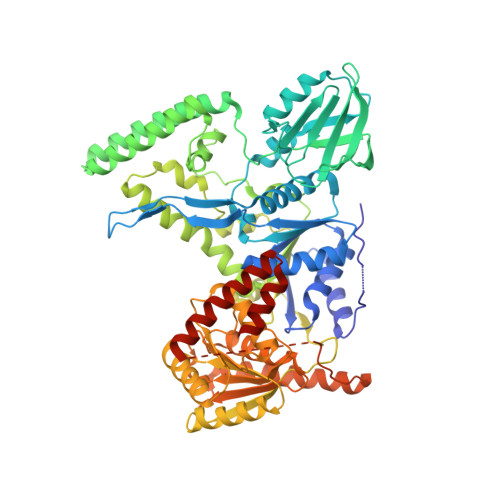
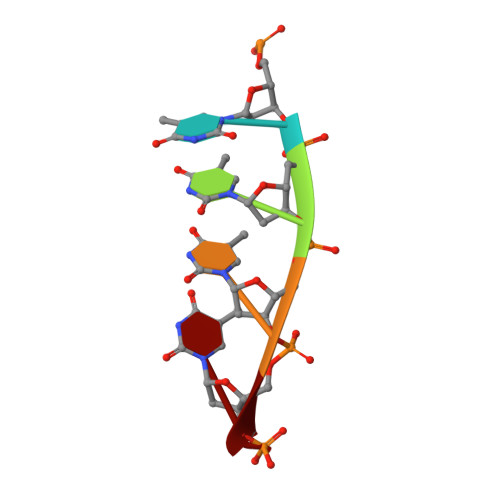
Damage detection by the UvrABC pathway: crystal structure of UvrB bound to fluorescein-adducted DNA
Waters, T.R., Eryilmaz, J., Geddes, S., Barrett, T.E.(2006) FEBS Lett 580: 6423-6427
- PubMed: 17097086
- DOI: https://doi.org/10.1016/j.febslet.2006.10.051
- Primary Citation of Related Structures:
2NMV - PubMed Abstract:
UvrB is the damage recognition element of the highly conserved UvrABC pathway that functions in the removal of bulky DNA adducts. Pivotal to this is the formation of a damage detection complex that relies on the ability of UvrB to locate and sequester diverse lesions. Whilst structures of UvrB bound to DNA have recently been reported, none address the issue of lesion recognition. Here, we describe the crystal structure of UvrB bound to a pentanucleotide containing a single fluorescein-adducted thymine that reveals a unique mechanism for damage detection entirely dependent on the exclusion of lesions larger than an undamaged nucleotide.
- The School of Crystallography and the Institute for Structural Molecular Biology, Birkbeck College, Malet Street, London WC1E 7HX, United Kingdom.
Organizational Affiliation: