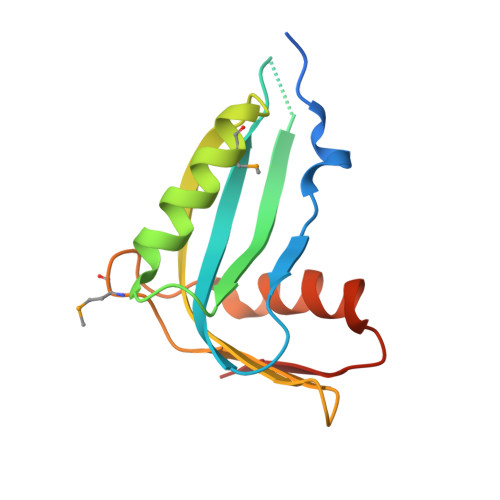
Crystal structure of human phosphohistidine phosphatase. Northeast Structural Genomics Consortium target HR1409
Kuzin, A.P., Abashidze, M., Forouhar, F., Seetharaman, J., Kent, C., Fang, Y., Cunningham, K., Conover, K., Ma, L.C., Xiao, R., Acton, T., Montelione, G., Tong, L., Hunt, J.F.To be published.