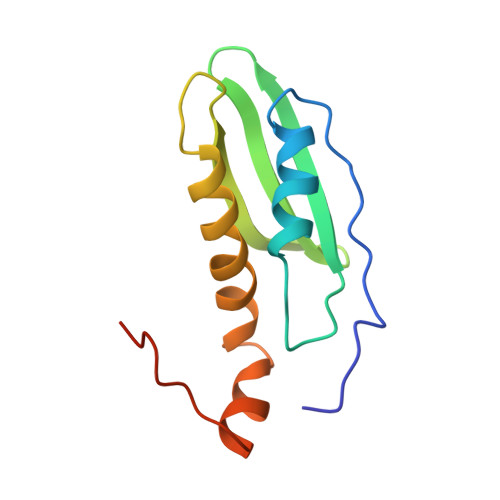
Solution NMR structure of CsgE: Structural insights into a chaperone and regulator protein important for functional amyloid formation.
Shu, Q., Krezel, A.M., Cusumano, Z.T., Pinkner, J.S., Klein, R., Hultgren, S.J., Frieden, C.(2016) Proc Natl Acad Sci U S A 113: 7130-7135
- PubMed: 27298344
- DOI: https://doi.org/10.1073/pnas.1607222113
- Primary Citation of Related Structures:
2NA4 - PubMed Abstract:
Curli, consisting primarily of major structural subunit CsgA, are functional amyloids produced on the surface of Escherichia coli, as well as many other enteric bacteria, and are involved in cell colonization and biofilm formation. CsgE is a periplasmic accessory protein that plays a crucial role in curli biogenesis. CsgE binds to both CsgA and the nonameric pore protein CsgG. The CsgG-CsgE complex is the curli secretion channel and is essential for the formation of the curli fibril in vivo. To better understand the role of CsgE in curli formation, we have determined the solution NMR structure of a double mutant of CsgE (W48A/F79A) that appears to be similar to the wild-type (WT) protein in overall structure and function but does not form mixed oligomers at NMR concentrations similar to the WT. The well-converged structure of this mutant has a core scaffold composed of a layer of two α-helices and a layer of three-stranded antiparallel β-sheet with flexible N and C termini. The structure of CsgE fits well into the cryoelectron microscopy density map of the CsgG-CsgE complex. We highlight a striking feature of the electrostatic potential surface in CsgE structure and present an assembly model of the CsgG-CsgE complex. We suggest a structural mechanism of the interaction between CsgE and CsgA. Understanding curli formation can provide the information necessary to develop treatments and therapeutic agents for biofilm-related infections and may benefit the prevention and treatment of amyloid diseases. CsgE could establish a paradigm for the regulation of amyloidogenesis because of its unique role in curli formation.
- Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. Louis, MO 63110;
Organizational Affiliation: