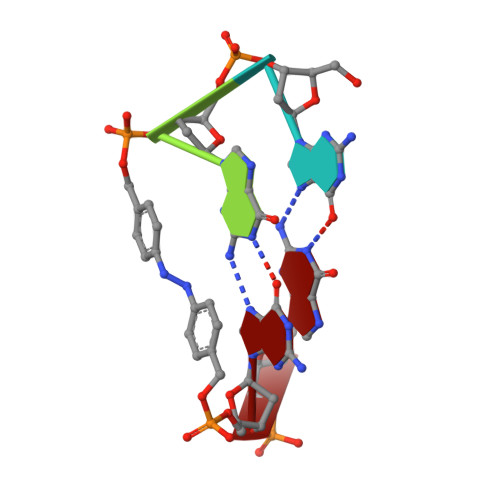
Photoresponsive Formation of an Intermolecular Minimal G-Quadruplex Motif.
Thevarpadam, J., Bessi, I., Binas, O., Goncalves, D.P., Slavov, C., Jonker, H.R., Richter, C., Wachtveitl, J., Schwalbe, H., Heckel, A.(2016) Angew Chem Int Ed Engl 55: 2738-2742
- PubMed: 26805928
- DOI: https://doi.org/10.1002/anie.201510269
- Primary Citation of Related Structures:
2N9Q - PubMed Abstract:
The ability of three different bifunctional azobenzene linkers to enable the photoreversible formation of a defined intermolecular two-tetrad G-quadruplex upon UV/Vis irradiation was investigated. Circular dichroism and NMR spectroscopic data showed the formation of G-quadruplexes with K(+) ions at room temperature in all three cases with the corresponding azobenzene linker in an E conformation. However, only the para-para-substituted azobenzene derivative enables photoswitching between a nonpolymorphic, stacked, tetramolecular G-quadruplex and an unstructured state after E-Z isomerization.
- Goethe University Frankfurt, Institute for Organic Chemistry and Chemical Biology, Buchmann Institute for Molecular Life Sciences, Max-von-Laue-Strasse 9, 60438, Frankfurt, Germany.
Organizational Affiliation: