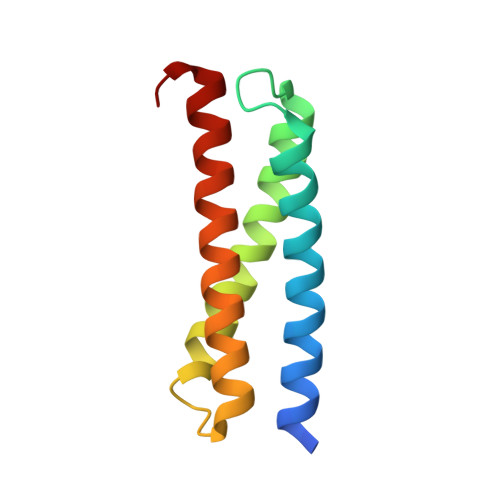
Structure of the GAT domain of the endosomal adapter protein Tom1.
Xiao, S., Ellena, J.F., Armstrong, G.S., Capelluto, D.G.(2016) Data Brief 7: 344-348
- PubMed: 26977434
- DOI: https://doi.org/10.1016/j.dib.2016.02.042
- Primary Citation of Related Structures:
2N9D - PubMed Abstract:
Cellular homeostasis requires correct delivery of cell-surface receptor proteins (cargo) to their target subcellular compartments. The adapter proteins Tom1 and Tollip are involved in sorting of ubiquitinated cargo in endosomal compartments. Recruitment of Tom1 to the endosomal compartments is mediated by its GAT domain's association to Tollip's Tom1-binding domain (TBD). In this data article, we report the solution NMR-derived structure of the Tom1 GAT domain. The estimated protein structure exhibits a bundle of three helical elements. We compare the Tom1 GAT structure with those structures corresponding to the Tollip TBD- and ubiquitin-bound states.
- Protein Signaling Domains Laboratory, Department of Biological Sciences, Biocomplexity Institute, Virginia Tech, Blacksburg, VA 24061, USA; School of Materials and Metallurgy, Inner Mongolia University of Science and Technology, PR China.
Organizational Affiliation: