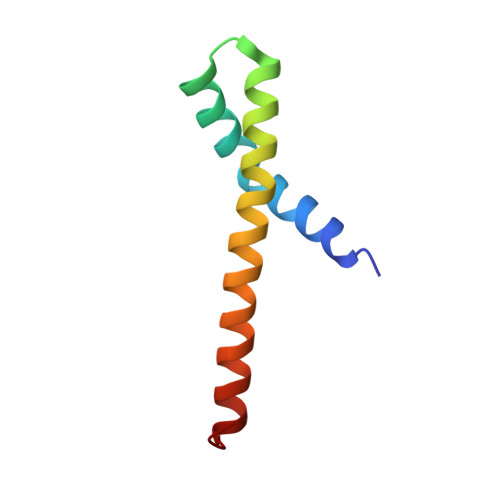
Structural insights and in vitro reconstitution of membrane targeting and activation of human PI4KB by the ACBD3 protein.
Klima, M., Toth, D.J., Hexnerova, R., Baumlova, A., Chalupska, D., Tykvart, J., Rezabkova, L., Sengupta, N., Man, P., Dubankova, A., Humpolickova, J., Nencka, R., Veverka, V., Balla, T., Boura, E.(2016) Sci Rep 6: 23641-23641
- PubMed: 27009356
- DOI: https://doi.org/10.1038/srep23641
- Primary Citation of Related Structures:
2N72, 2N73 - PubMed Abstract:
Phosphatidylinositol 4-kinase beta (PI4KB) is one of four human PI4K enzymes that generate phosphatidylinositol 4-phosphate (PI4P), a minor but essential regulatory lipid found in all eukaryotic cells. To convert their lipid substrates, PI4Ks must be recruited to the correct membrane compartment. PI4KB is critical for the maintenance of the Golgi and trans Golgi network (TGN) PI4P pools, however, the actual targeting mechanism of PI4KB to the Golgi and TGN membranes is unknown. Here, we present an NMR structure of the complex of PI4KB and its interacting partner, Golgi adaptor protein acyl-coenzyme A binding domain containing protein 3 (ACBD3). We show that ACBD3 is capable of recruiting PI4KB to membranes both in vitro and in vivo, and that membrane recruitment of PI4KB by ACBD3 increases its enzymatic activity and that the ACBD3:PI4KB complex formation is essential for proper function of the Golgi.
- Institute of Organic Chemistry and Biochemistry AS CR, v.v.i., Flemingovo nam. 2., 16610 Prague 6, Czech Republic.
Organizational Affiliation: